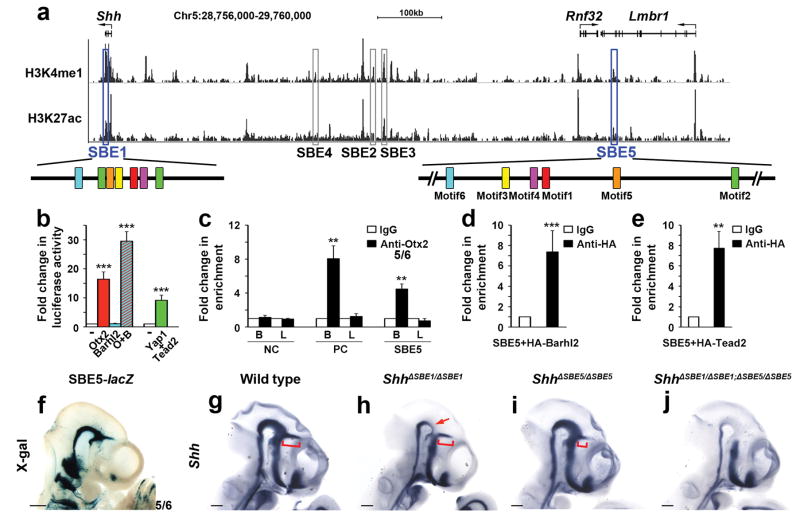
Figure 3. Identification of SBE5 as a functional SBE1 homologue.
(a) UCSC genome browser view (NCBI37/mm9) of a 1 Mb interval upstream of Shh showing ChIP-seq signal enrichment for chromatin marks (H3K27ac and H3K4me1) associated with enhancer activity in whole brain (E14.5)34. The location of SBE1 and SBE5 related peaks (outlined in blue), other Shh CNS enhancer related peaks (outlined in grey) and the shuffled arrangement of their motifs (colored boxes) are displayed. (b) Activation of an SBE5-luciferase reporter construct by members of the SBE1 transcription factor collective. (c–e) ChIP-QPCR shows SBE5 enrichment in Otx2 bound chromatin from embryonic brain (B) but not limb bud (L), as well as Barhl2 and Tead2 bound chromatin from tissue culture cells. Negative (NC) and Positive (PC) control primers in (c) amplify sequence upstream of SBE1 and within an Emx2 forebrain enhancer, respectively (**P<0.01, ***P<0.001, Student’s t-test). Error bars represent s.d.m. Each bar (b–e) represents the average of at least three experiments performed in triplicate. (f) X-gal staining of a transgenic embryo (E10.5) expressing SBE5-lacZ in a similar pattern to SBE1 (compare with Fig. 2a). (g–j) Whole mount in situ hybridization for Shh on control and SBE1/SBE5 single and double mutant embryos (E10.5). ShhΔSBE1/ΔSBE1embryos show reduced Shh expression in the ventral midbrain and caudal diencephalon (red arrow). ShhΔSBE5/ΔSBE5 embryos display a partial truncation in Shh zli expression (red bracket). ShhΔSBE1/ΔSBE1; ΔSBE5/ΔSBE5 embryos are devoid of Shh in the entire SBE1 domain. Scale bars: 0.5mm.