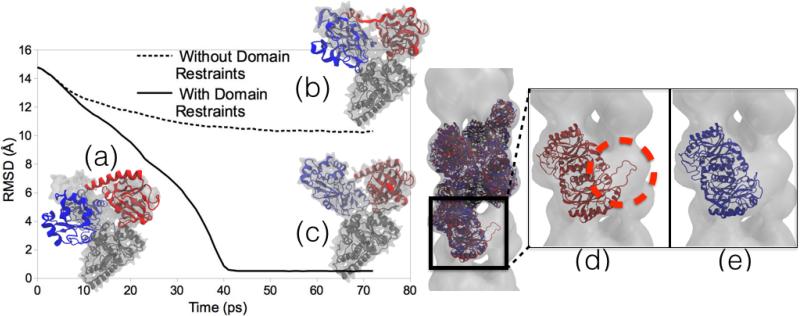
Figure 2.
Domain and symmetry restraints for MDFF. Left Image: RMSD plots of MDFF trajectories and representative structures. (a) Initial docking of Acetyl-CoA Synthase structure into cryo-EM density. (b) A structure obtained from MDFF without using domain restraints applied to the two domains (red and blue) to keep the local structure intact is distorted and improperly fit to the density. (c) Result of MDFF with application of domain restraints. The graph shows the RMSD of the structures to a known validation model, with the non-domain-restrained simulation exhibiting a much worse fit. Right Image: A two-turn helix model of a nitrilase from R. rhodochrous J1 [72, 57] fit to a larger cryo-EM density. (d) Final fitted structure from MDFF without using symmetry restraints shows the edge-distortion effect of a subunit being pulled into empty adjacent density (circled in red). (e) Final fitted structure from MDFF using symmetry restraints avoids the edge-distortion effect by restraining the subunits to an average structure.