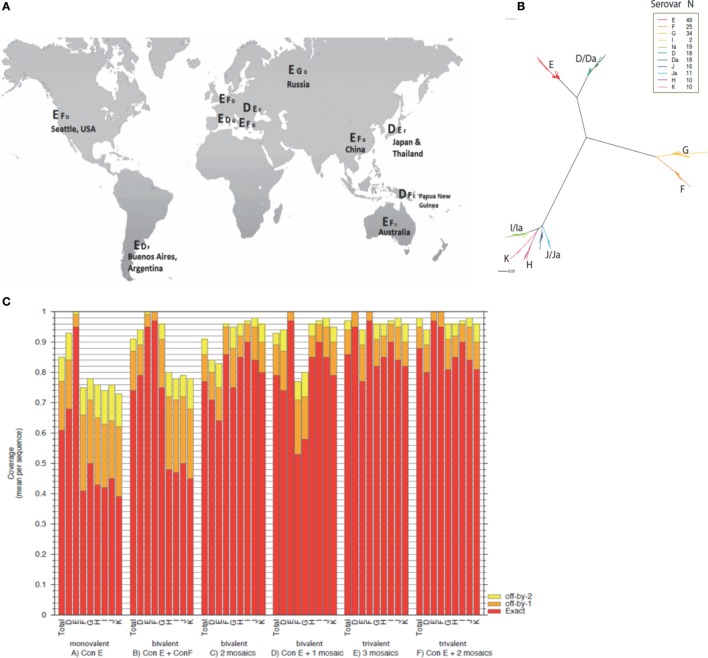
Figure 1.
Global C. trachomatis serovar prevalence, phylogeny, and theoretical epitope coverage of consensus and mosaic MOMP antigens. (A) The serovar prevalence of C. trachomatis worldwide compiled from a literature review and represented in descending prevalence at global locations. (B) A phylogenetic maximum likelihood tree based on the ompA sequence alignments derived from Ref. (24) was created using FastTree and the graphic generated by Rainbow Tree. (C) Potential epitope coverage against all serovars (total) and individual serovars (serovars D–K) were analyzed for a monovalent Con E antigen, Con E and Con F antigens, two mosaic antigens, a Con E antigen and a mosaic antigen, three mosaic antigens, and a Con E antigen with two additional mosaic antigens using EPICOVER. Mean 9-mer coverage presented against individual and total combined serovars D–K, with exact (red), off-by-1 (orange), and off-by-2 (yellow) epitope matching.