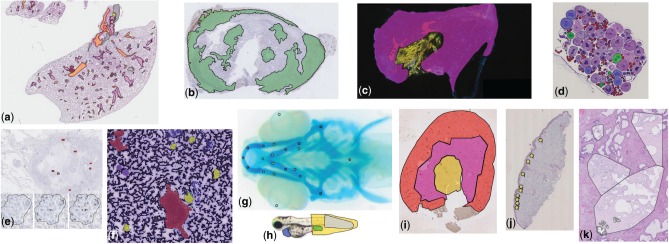
Fig. 3.
Examples of annotations created using Cytomine in images from various research fields (see Section 3 for additional details): (a) Delineation of tissue components in H&E images in mice lung cancer research (D.Cataldo’s lab), (b) Tumoral areas in HDAB images in mice lung cancer research (P. Martinive’s lab), (c) Area quantification in immunofluorescent mouse ear sponge assays in tumor angiogenesis (C. Gilles’ lab), (d) Counting of oocytes in H&E images in Chondrostoma nasus sexual maturation research (V. Gennotte’s lab), (e) mRNA expression quantification through in situ hybridization assays in human breast cancer research (C.Josse’s lab), (f) Cell types in fine-needle aspiration cytology in human thyroid (I. Salmon’s lab), (g) Landmarks in Danio rerio embryo development (M. Muller’s lab), (h) Phenotypes in Danio rerio toxicology research (M.Muller’s lab), (i) Region delineation and cell counting in immunohistochemistry images in renal ischemia/reperfusion research (F.Jouret’s lab), (j) Cell scoring in immunohistochemistry images in melanoma microenvironment research (P.Quatresooz’s lab), (k) Nucleus counting in H&E images in human breast cancer research (E. De Pauw’s lab)