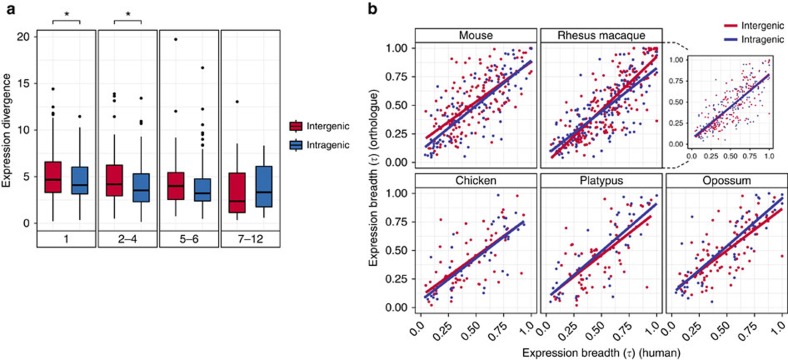
Figure 4. Interspecies analyses of miRNA expression.
(a) Expression divergence between human miRNAs and their orthologues in five species (chicken, platypus, opossum, mouse and rhesus macaque). The expression divergence was calculated by means of Euclidean distances of expression levels across five tissues (brain, cerebellum, heart, kidney and testis) for each pair of orthologues. Box plots represent the distribution of pooled distances for each pair according to miRNA age class. Significant differences were assessed by Mann–Whitney U-tests (*P<0.02). (b) Spearman's correlations between expression breadth of human miRNAs and their orthologues with respect to their genomic context (inter- and intragenic). Correlations are as follows: Chicken: intergenic: ρ=0.70, intragenic: ρ=0.78; Platypus: intergenic: ρ=0.68, intragenic: ρ=0.83; Opossum: intergenic: ρ=0.76, intragenic: ρ=0.85; Mouse: intergenic: ρ=0.69, intragenic: ρ=0.81; Rhesus macaque: intergenic: ρ=0.86, intragenic: ρ=0.79; Rhesus macaque excluding miRNAs of age 7 (shown in detail): intergenic: ρ=0.79, intragenic: ρ=0.81 (all P<1.0 × 10−9). To assess the significance of the difference between two correlation coefficients, we used the Fisher z transformation. Significant differences between inter- and intragenic correlations were observed for Platypus (one-tailed P=0.02), Opossum (one-tailed P=0.05) and Mouse (one-tailed P=0.01). A minimum of 1 c.p.m. (counts per million) in at least one tissue was adopted as expression threshold for both analyses (a,b).