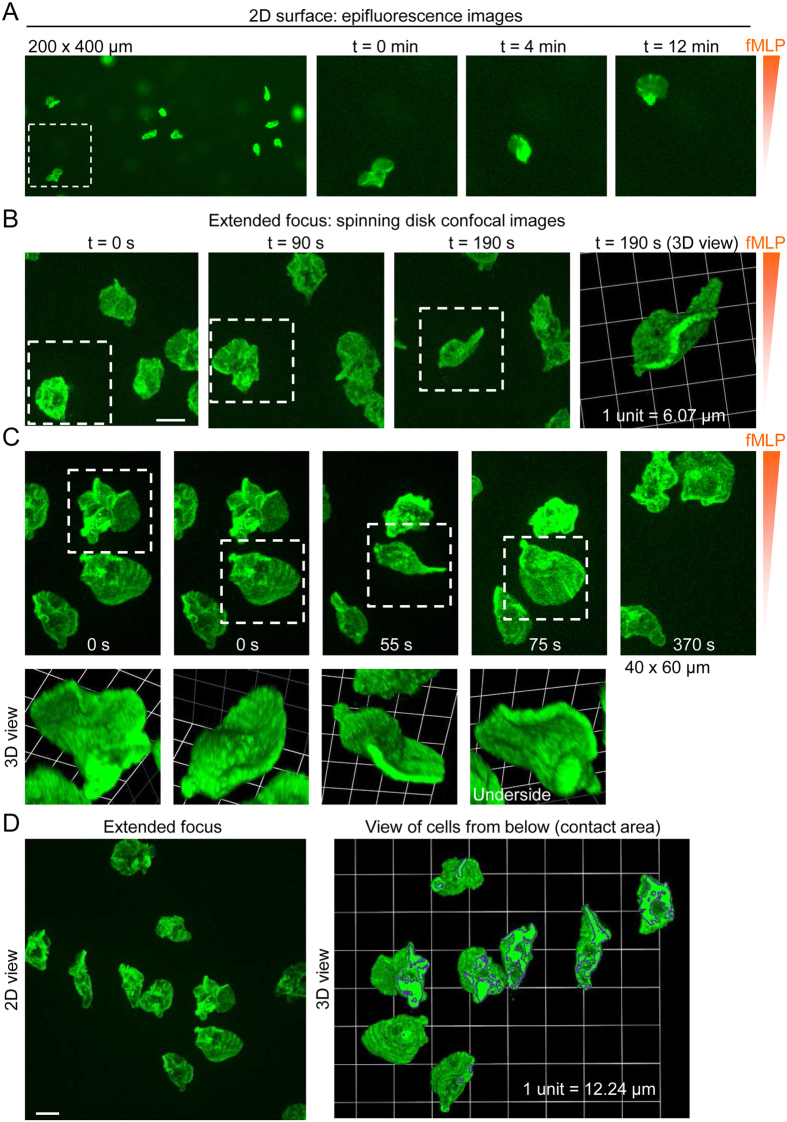
Figure 3. 3D morphology of human monocytes migrating on a 2D substrate.
(A) Snapshot (200 × 400 μm) and enlarged (cropped) time-lapse images (obtained by epi-fluorescence imaging) of human monocytes in a chemotactic fMLP gradient. (B) Projected (extended focus) fluorescence images, obtained by time-lapse spinning disk confocal microscopy, of monocytes migrating in an fMLP gradient (scale bar, 10 μm). The white squares track the same cell at different time points, and a 3D view corresponding to t = 190 s is shown on the right. (C) Projected (extended focus) fluorescence images (40 × 60 μm; upper panel), obtained by time-lapse spinning disk confocal microscopy, and corresponding reconstruction of 3D morphology (lower panel). (D) View of human monocytes from above (2D extended focus view; scale bar, 10 μm) and from below (after rotating the image 180° on the x-axis), showing the cell-substrate contact areas, highlighted by dark boundaries.