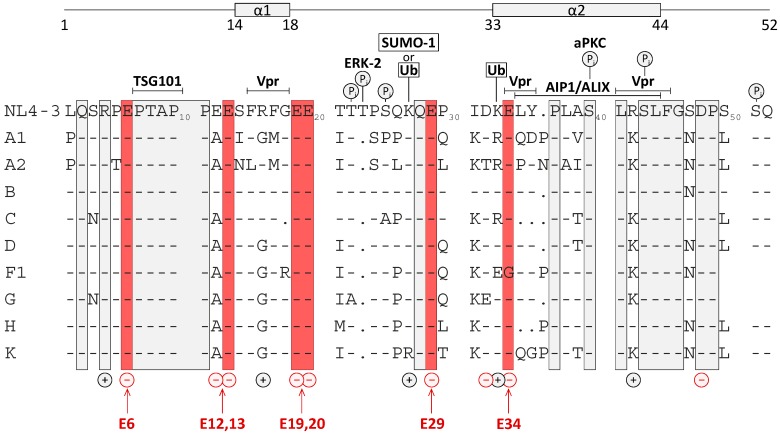
Figure 1.
Amino acid sequence of p6 derived from the isolate HIV-1NL4-3 with previously identified structural and functional domains [50], charge distribution and sites of posttranslational modifications. Delineated in red are conserved Glu residues at positions 6, 13, 19, 20, 29, and 34. Positively and negatively charged residues, previously identified phosphorylation sites for ERK-2 and aPKC [35,36], attachment sites for ubiquitin [21] and SUMO-1 [22], binding domains for Tsg101 [8,9,10,34,51], ALIX [12], and Vpr [49] and structural domains identified by NMR [50] are indicated. Consensus sequences of p6 proteins derived from the M-group viruses [52] were aligned and conserved amino acids are boxed in grey.