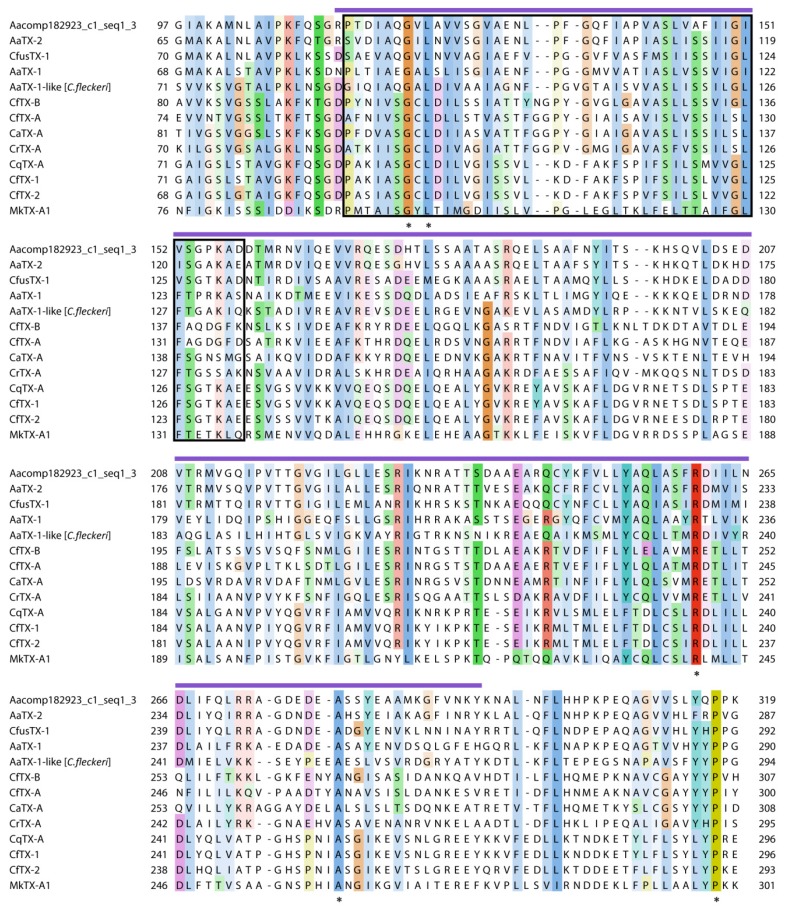
Figure 5.
Partial multiple protein sequence alignment of CfusTX-1 and related jellyfish toxins highlighting the regions of highest sequence similarity. Sequences were aligned using MUSCLE and visualized using Jalview. Amino acid residue shading is based on the Clustal protein colour scheme, with color intensity increasing as residue conservation increases from 25% to 100%. Identical residues are indicated with an asterisk. Dashes represent gaps introduced for better alignment. A predicted transmembrane spanning region (TSR1) that is common among the jellyfish toxins is indicated with a black outline. A purple line above the alignment corresponds to a predicted δ-endotoxin, N-terminal-like domain. References for Aacomp182923_c1_seq1_3 [24] and AaTX-1-like [26].