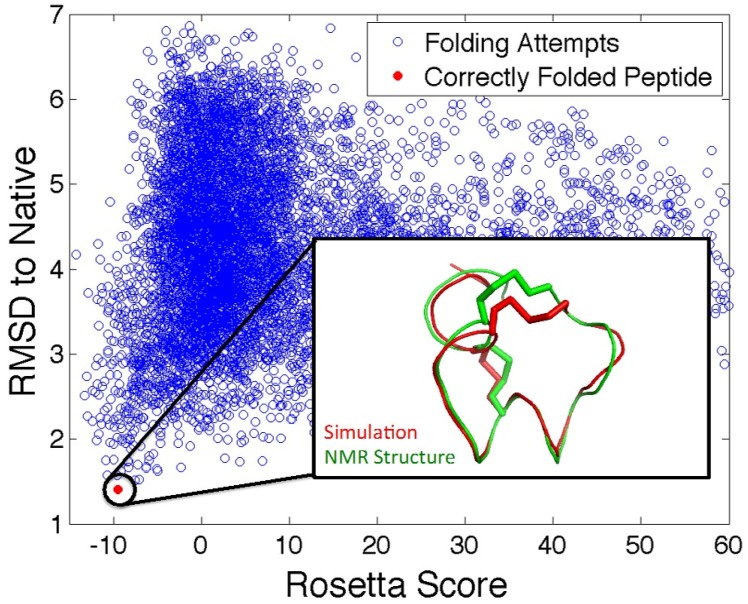
Figure 6.
Predicting 3D structure of venom peptides. Scatter plot representation of Rosetta scores for each of the 10,000 attempts to fold α-GID conotoxin from its amino acid sequence. Blue circles represent each folding attempt and the red circle represents a folding simulation that resulted in the correct structure. Inset: comparison of α-GID NMR structure (green) and Rosetta structure prediction (red). Rosetta ab initio folding protocol was used to predict structure and scores were calculated as the Root-Mean-Square Deviation (RMSD) to the NMR structure of α-GID.