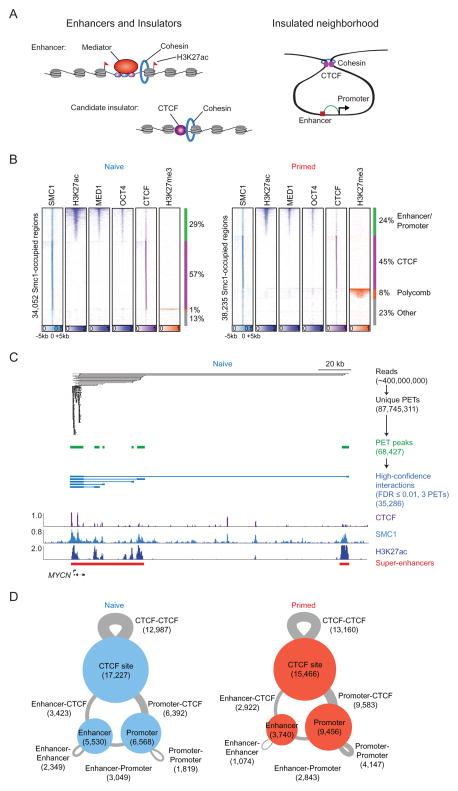
Figure 1. Components of 3D regulatory landscape.
(A) Enhancers and insulators (left panel). Enhancers are occupied by transcription factors, mediator and cohesin, and their associated nucleosomes are marked by H3K27ac. Candidate insulators are occupied by CTCF and cohesin. Model of insulated neighborhoods formed by cohesin-associated CTCF-CTCF interactions, within which enhancers loop to promoters of target genes (right panel).
(B) Heatmap representation of ChIP-seq data for H3K27ac, MED1, OCT4, CTCF and H3K27me3 at SMC1-occupied regions in naive (left panel) and primed (right panel) hESCs. Read density is displayed within a 10 kb window and color scale intensities are shown in rpm/bp. Cohesin occupies three classes of sites: enhancer-promoter sites, Polycomb-occupied sites, and CTCF-occupied sites.
(C) Cohesin (SMC1) ChIA-PET data analysis at the MYCN locus in naive hESCs. The algorithm used to identify paired-end tags (PETs) is described in Extended Experimental Procedures. PETs and interactions involving enhancers and promoters within the window are displayed at each step in the analysis pipeline. Binding profiles for CTCF, SMC1 and H3K27ac are displayed at the bottom.
(D) High-confidence cohesin-associated interaction maps in naive (left panel) and primed (right panel) hESCs. CTCF binding sites, enhancers and promoters involved in cohesin-associated interactions are indicated as circles, and the size of circles correspond to the number of sites. The interactions between two regions are indicated as gray lines, and the size of lines correspond to the number of interactions.