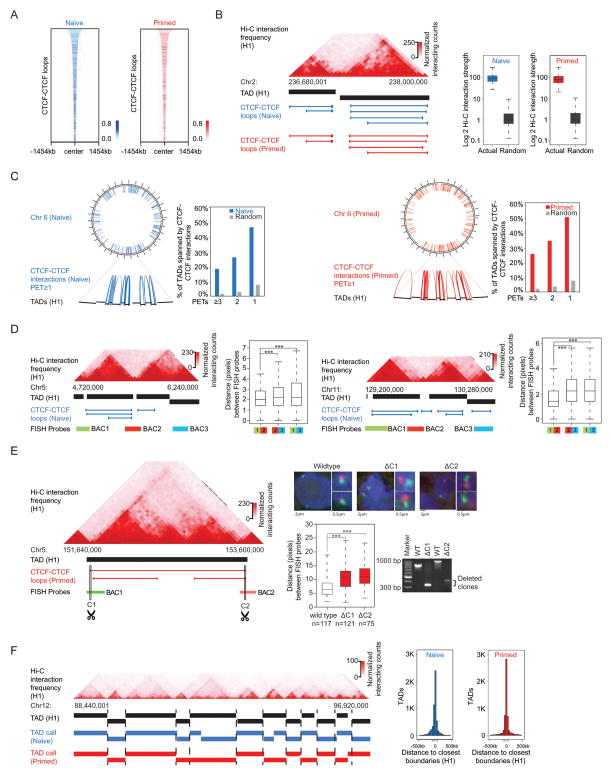
Figure 2. CTCF-CTCF loops underlie much of TAD structure.
(A) Heatmap of cohesin-associated CTCF-CTCF loops showing that these loops in naive hESCs are largely preserved in primed hESCs. The 9,344 CTCF-CTCF loops that define the putative insulated neighborhoods in naive hESCs were ranked by size and shown. The color bar indicates normalized PET-signal at these CTCF-CTCF loops.
(B) TAD heat map of interaction frequencies and CTCF-CTCF loops that define the putative insulated neighborhoods. Normalized Hi-C interaction frequencies in H1 hESCs are displayed in a two-dimensional heat map (Dixon et al., 2015) with the TADs indicated as black bars. Shared CTCF-CTCF loops are indicated as blue lines (naive) and red lines (primed). A correlation analysis between Hi-C interaction frequency (H1 hESCs) and CTCF-CTCF loops in naive and primed hESCs is displayed to the right in a box plot; randomly generated TADs were used as the background control.
(C) CTCF-CTCF loops span many TADs identified using Hi-C data in H1 hESCs. Chromosome 6 is displayed as a circos plot in both naive and primed hESCs, with zoomed in regions below. CTCF-CTCF loops (≥1 PETs) are indicated as blue arcs (naive) and red arcs (primed). The bar graphs show percentages of TADs spanned by CTCF-CTCF loops when various confidence thresholds (1, 2, ≥3 PETs) were used. Random shuffling of TAD locations (100 iterations) serve as the background control.
(D) Physical distance between TAD borders is shorter than an equidistant control locus. The Hi-C interaction heatmaps, TADs and CTCF-CTCF loops were shown the same as (B). The green, red and blue bars indicate the location of BAC probes used for DNA FISH at each locus. Box plots of minimal normalized distances between pairs of loci generated from >1500 FISH probe spots per condition are displayed with the corresponding probe pairs labeled below. The stars indicate significance using the Mann-Whitney test (***P<10−28). Images were obtained using a 40X objective.
(E) Measurement of DNA proximity by 3D DNA FISH before and after deletions of CTCF binding sites at either end of a TAD-spanning CTCF-CTCF loop. The Hi-C interaction heatmaps, TADs and CTCF-CTCF loops were shown the same as (B). The green and red bars indicate the location of BAC probes used for DNA FISH. The scissor-marked regions (C1, C2) were deleted by CRISPR-mediated deletion. Examples of two color DNA FISH images are shown in the right panel, the quantification of distance between green and red probes are displayed with bar graphs shown below. The stars indicate significance using the Mann-Whitney test (***P<10−13). Images were obtained using a 100X objective; the n indicates the number of alleles quantified for each sample. The genotyping PCR data are displayed at the bottom right.
(F) Cohesin ChIA-PET data can be used to discover TADs. A comparison of TADs derived with the same algorithm from Hi-C data (Dixon et al., 2015) and cohesin ChIA-PET data for a portion of chromosome 12 (left panel). A global analysis indicates that the cohesin ChIA-PET and Hi-C derived TAD boundaries are close (right panel).