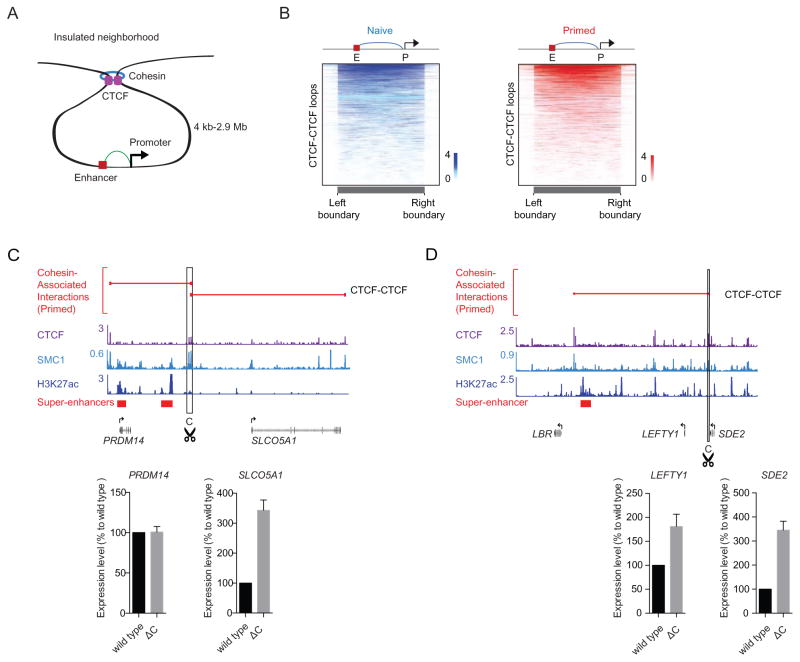
Figure 3. Putative insulated neighborhoods in hESCs.
(A) Schematic of insulated neighborhood.
(B) Enhancer-promoter interactions occur predominantly within CTCF-CTCF loops that define putative insulated neighborhoods in hESCs. The color bar indicates the number of enhancer-promoter interactions spanning the genomic location.
(C) and (D) CRISPR-mediated deletion of CTCF sites at two loci (PRDM14 locus (C) and LEFTY1 locus (D)). The top of each panel shows a subset of CTCF-CTCF loops depicted as red lines and binding profiles for CTCF, cohesin (SMC1), and H3K27ac in primed hESCs at the respective loci. A subset of genes present in these loops is shown for simplicity. The super-enhancers are indicated as red bars. The bottom of each panel shows RT-qPCR results for the gene expression levels of the indicated genes in wild type and cells with deleted CTCF sites. Error bars were generated from at least three replicates.
See also Figure S3