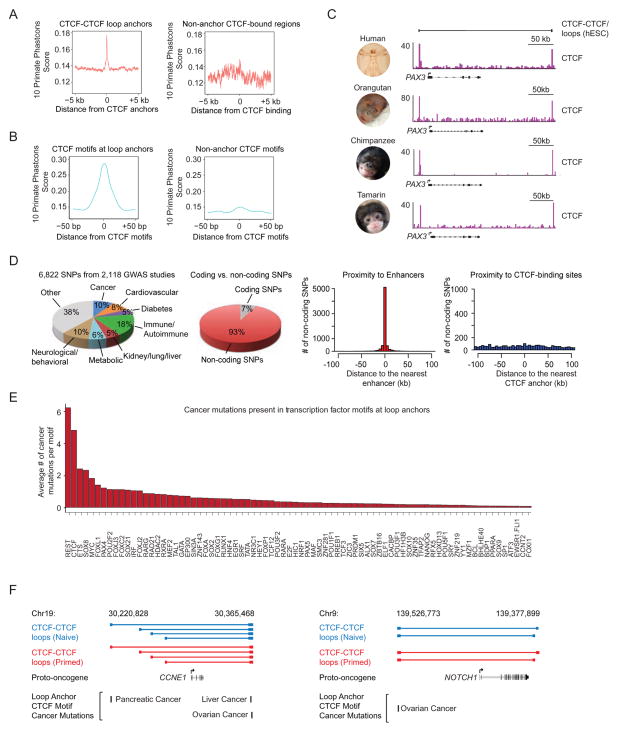
Figure 6. Conservation of 3D structure and associations with disease.
(A) and (B) DNA sequence in anchor regions (A) and the CTCF DNA sequence motif (B) of CTCF-CTCF loops in hESCs is more conserved in primates than DNA sequence in hESC regions bound by CTCF that do not serve as loop anchors.
(C) A CTCF-CTCF loop containing the PAX3 gene in human and ChIP-seq gene tracks showing conserved binding of CTCF at this locus in Human, Orangutan, Chimpanzee and Tamarin genomes (Schwalie et al., 2013).
(D) Catalog of SNPs linked to phenotypic traits and diseases in genome-wide association studies (GWAS) and SNP association with enhancer and CTCF anchor regions in hESCs. Pie chart showing percentage of SNPs associated with the highlighted classes of traits and diseases (Left). Distribution of trait-associated SNPs in coding and noncoding regions of the genome (Middle Left). Location of all noncoding trait-associated SNPs relative to all enhancers identified in 86 human cell and tissue samples. x axis reflects binned distances of each SNP to the nearest enhancer. SNPs located within enhancers are assigned to the 0 bin (Middle Right). Location of all noncoding trait-associated SNPs relative to CTCF binding sites in loop anchor regions (Right).
(E) Cancer mutations in transcription factor motifs at hESC CTCF-CTCF loop anchors.
(F) Cancer mutations found at CTCF motifs at the anchors of CTCF-CTCF loops in hESCs that contain the proto-oncogenes CCNE1 and NOTCH1. Blue (naive) and red (primed) CTCF-CTCF loops with mutations within the CTCF motifs in their anchors are displayed above the proto-oncogene contained within these loops. Below, mutations from the International Cancer Genome Consortium are displayed along with the cancers from which these were sequenced.