Fig. 5.
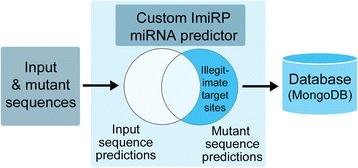
The target site prediction module. Predicted miRNA target sites within input and mutant sequences are identified using a custom component that identifies predicted target sites based on complementarity to the miRNA 5’ end. FASTA-format miRNA sequence data was collected from miRBase version 21 [11]. Information about predicted sites present in each mutant sequence that are absent from the input sequence are stored in a database for analysis
