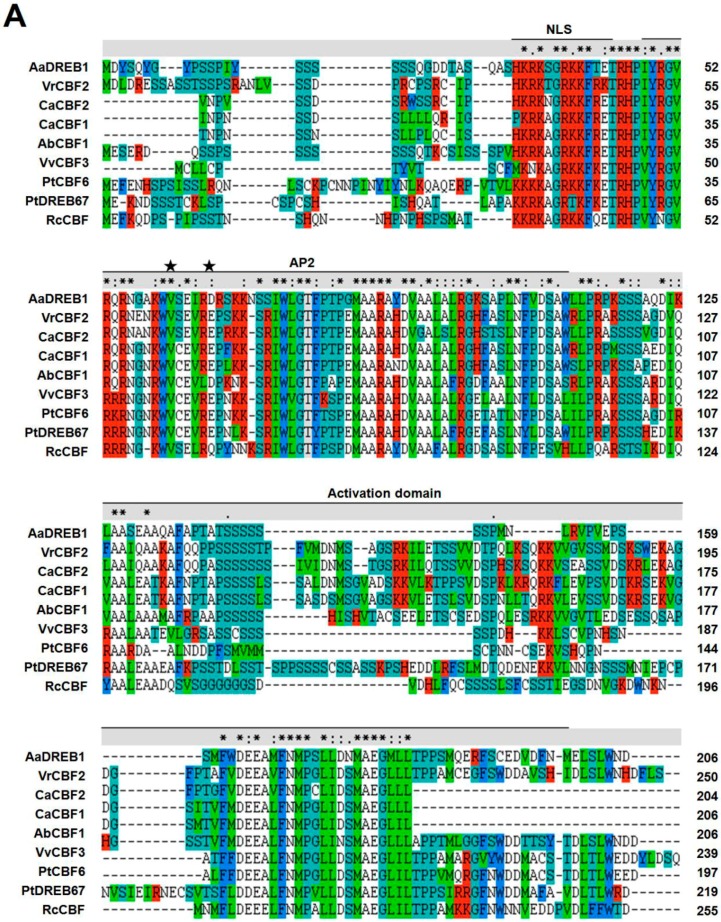
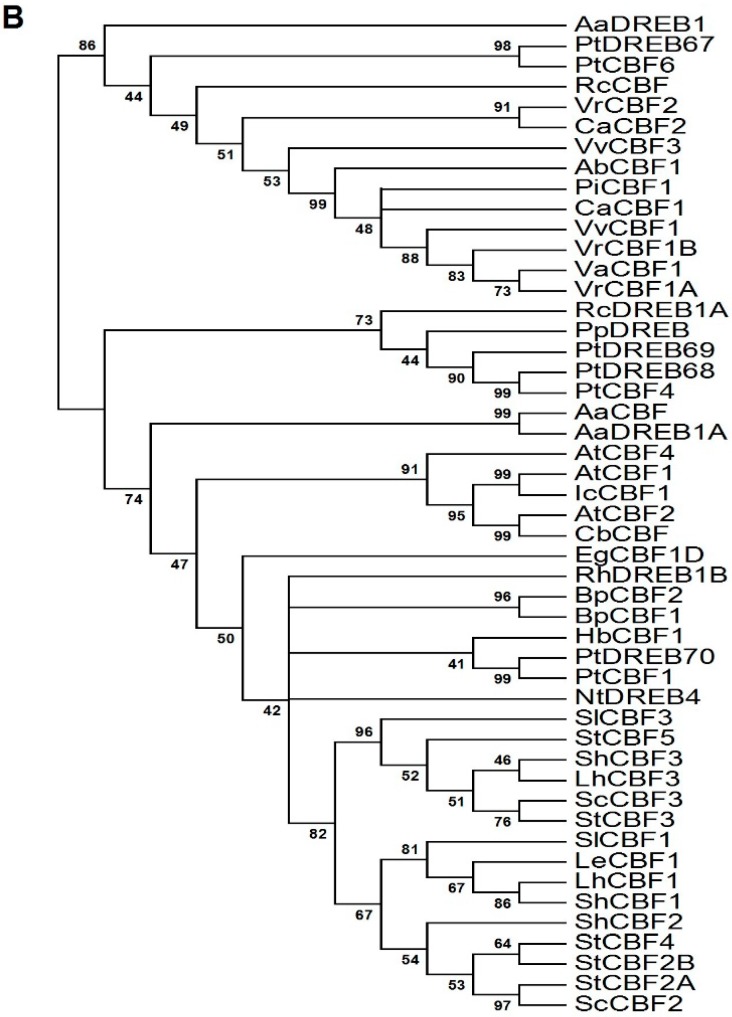
Figure 1.
Comparison of the deduced amino acid sequence of Adonis amurensis dehydration-responsive element binding protein 1 (AaDREB1) and its homologs. Information of the DREB proteins of plants used here are in Supplementary Materials 1. (A) The sequences were aligned using CLUSTAL X. Gaps were introduced to optimize the alignment. The asterisks “*” indicate positions with a single, fully conserved residue; ‘‘:’’ indicates highly conserved positions; and “.” indicates more weakly conserved positions. Asterisks mark 14th and 19th amino acids of APETALA2/ethylene-responsive element-binding factor (AP2/ERF) domain. Dashes indicate gaps in the amino acid sequences. The column shows the score of the conservation positions. Lines above the alignment indicate the nuclear localization signal (NLS), the conserved AP2 domain, and the activation domain; identical and similar amino acids sequences are highlighted by red, deongaree, Cambridge blue and green shading respectively; (B) Phylogenetic analysis of Adonis amurensis and other plants’ DREB family proteins. Analysis, based on minimum evolution, was performed with full-length protein sequences using the AP2 transcription factor as an outgroup. DREB proteins were initially aligned using Clustal W and were used for phylogenetic analysis using MEGA version 4.1 software. The phylogenetic tree was constructed using the neighbor-joining method with 1000 bootstrap replications. Bootstrap percentages are shown at dendrogram branch points.