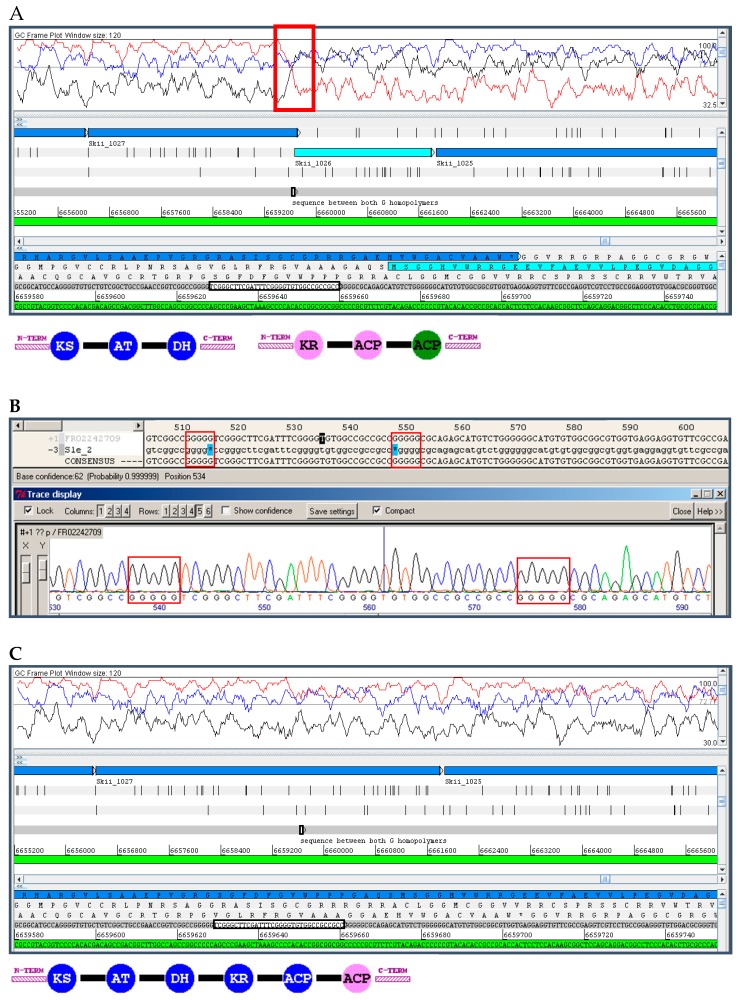
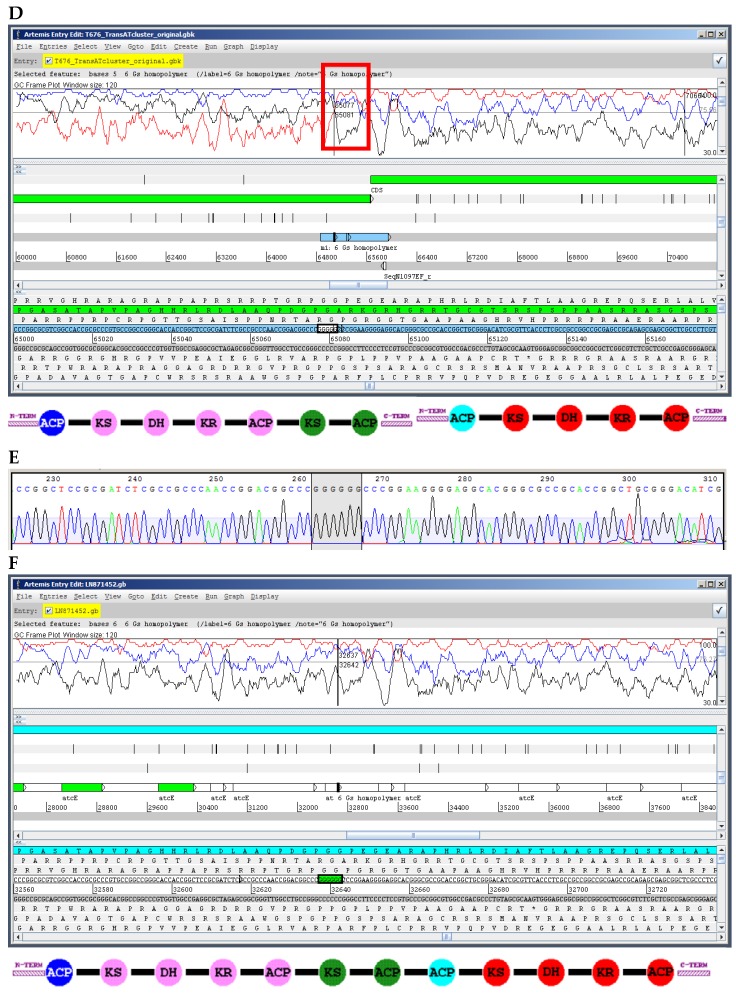
Figure 4.
Illustration of shifts in the reading frame caused by omission of one nucleotide in homopolymers of G or C in PacBio assemblies, and their effect on the modularity of PKS genes of the chaxamycin [43,55] (A–C) and anthracimycin [57] (D–F) gene clusters. A,D. Original PacBio sequence showing the frame-shift (red box) and the break in the modularity of the PKS domains; in A, the frame shift is obvious since it splits the module of PKS domains into two protein coding sequences; in D, while each PKS protein exhibits an appropriate modular domain structure (non-canonical domain arrangements are not unusual in trans-AT PKSs [57]) note that the 3’ end of the first protein coding sequence continues in the wrong reading frame which would correspond to a highly non-canonical codon usage. E. Sanger sequencing of the affected region showing the affected homopolymer; in the chaxamycin gene cluster these errors had been also corrected with Illumina reads [43]; in the anthracimycin example, the entire region in the cyan box (D) was sequenced with Sanger, and the only error found was the one shown in E. C,F. Organisation of the PKS gene and domain modularity after error correction. PKS domains identified with the SBSPKS server [63]: KS, ketosynthase; AT, acyltransferase; DH, dehydratase; KR, ketoreductase; ACP, acyl-carrier protein; Note that there is a methyltransferase domain in the last module that has not been identified by SBSPKS.