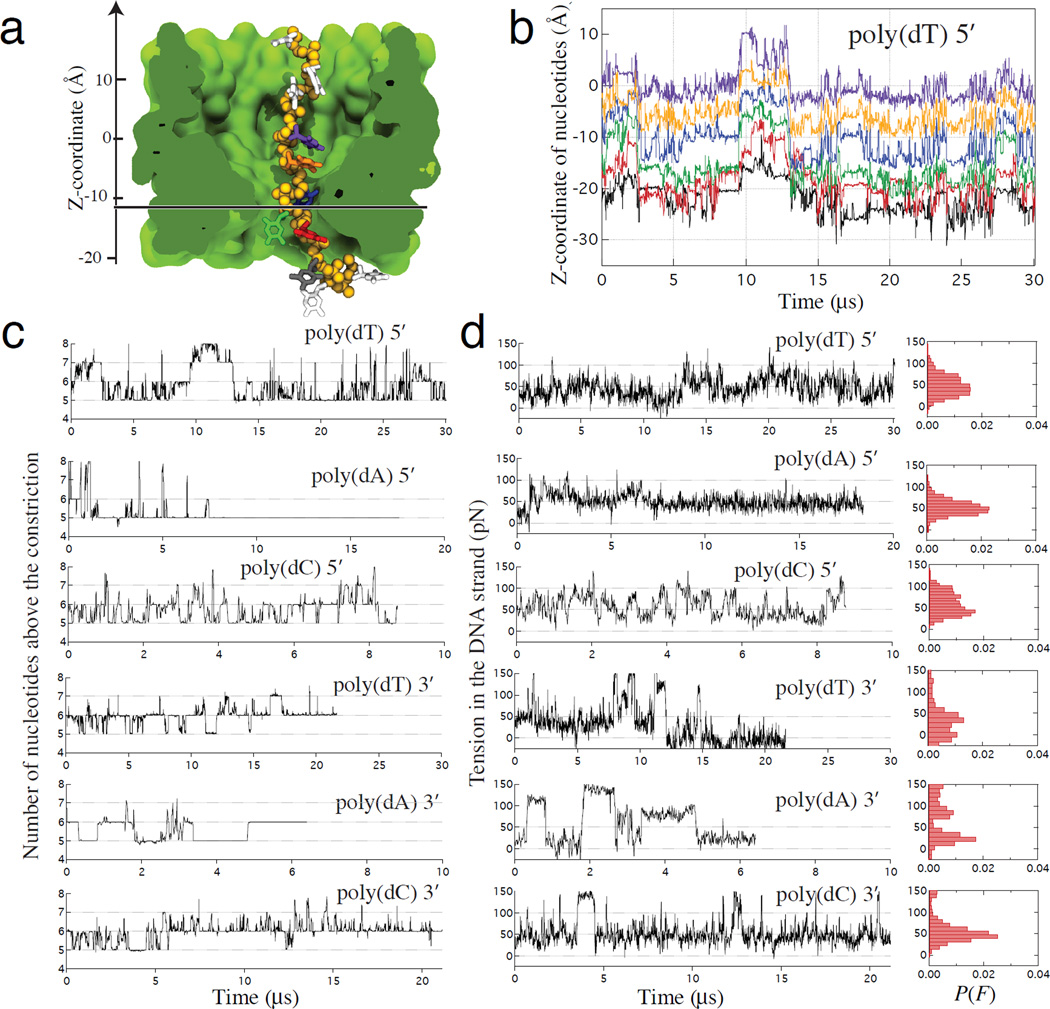
Figure 5. Stochastic displacement of nucleotides and the effective force.
(a) A typical simulation system containing MspA (green) and a DNA strand attached to a harmonic spring. Extension of the spring reports the effective force applied to DNA. The backbone of DNA is shown as yellow spheres, the DNA bases are individually colored. Lipids, water and ions are not shown. The horizontal line indicates the plane passing through the middle of the MspA constriction. (b) Centers of mass of DNA nucleotides versus simulation time in a representative MD trajectory. The color of the lines corresponds to the color of nucleotides shown in panel a. The z coordinate is defined in panel a. The dashed line indicates the location of the middle plane of the MspA constriction. For clarity, data are shown only for six nucleotides nearest to the constriction. (c) The number of nucleotides above the middle plane of the constriction versus simulation time for six homopolymer systems. (d) The force experienced by the harmonic spring attached to the uppermost nucleotide. Spontaneous, collective displacements of DNA nucleotides are associated with large fluctuations of the effective force applied to DNA. Normalized histograms summarize force distributions for each simulation system. In this figure, all data points represent 10-ns block averages of MD trajectories sampled at 100 ps. Because our simulations employed a reduced-length MspA system, the simulated values of the effective force can be up to 30% higher than the experimentally measured ones at the same transmembrane bias, Supplementary Note 4.