Abstract
Increasing evidence shows that long noncoding RNAs (lncRNAs) have important roles in the regulation of multiple cellular processes, including cell division, cell growth, and apoptosis, as well as cancer metastasis and neurological disease progression; however, the mechanism of how lncRNAs regulate these processes is not well established. In this study, we demonstrated that downregulating the expression of the lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) in breast cancer cells inhibited cell growth and induced cell apoptosis. In addition, the RNA-binding protein fused in sarcoma/translocated in liposarcoma (FUS/TLS) physically interacted with NEAT1, and reducing the expression of FUS/TLS also induced cell apoptosis. Multiple miRNAs were identified as regulators of NEAT1, but only overexpression of miR-548ar was able to decrease NEAT1 expression and promote apoptosis. These results indicate a novel interaction between NEAT1, miR-548ar-3p, and FUS and their role in the regulation of breast cancer cell apoptosis.
Keywords: lncRNA, NEAT1, miR-548ar-3p, FUS, cell apoptosis
Introduction
Advances in genomics over the past two decades have prompted researchers to reexamine noncoding RNAs (ncRNAs). As genomes of different mammalian species were sequenced, studies reported that protein-coding genes comprised only a tiny fraction of the total genomes, suggesting that the remaining genome likely had some function that remained to be explored. Efforts to explore this large and elusive portion of the human genome had yielded some 3,000 ncRNA genes with known functions, including most of the well- known small ncRNA, and some of the long noncoding RNAs (lncRNAs).1 Studies revealed lncRNAs as prominent regulators of several biological processes, including apoptosis,2 tumor development and progression,3 and metastases of cancer cells.4
Since lncRNAs represent an extensive and largely unexplored part of the genome,5 there has been considerable debate on the actual functions and processes by which lncRNAs interact with other systems. Two competing theories posit that lncRNAs function either as a sponge, absorbing microRNA and thereby regulating its target message RNA,6 or, alternatively, as chromatin regulators by binding to histones or other protein complexes and, thereby, regulating global gene transcription.7 While the sponge model has enjoyed comparatively stronger support, the chromatin model has received a substantial boost from investigation into nuclear paraspeckle assembly transcript 1 (NEAT1), which constitutes nuclear bodies known as paraspeckles that are found in all human cells, but whose function is not well understood. Recent studies found that NEAT1 plays a critical role in tissue development of the corpus luteum,8 placenta9 and mammary glands.10 Moreover, the expression of Neat1 is induced upon immune responses to viral infections11 and is also involved in tumorigenesis, including leukemia12 and prostate cancer.13 Neat1, spliced by serine/arginine-rich splicing protein, could regulate PARγ2, which is a pivotal molecule for adipogenesis.14 Collectively, these biological functions were theoretically postulated via regulation of transcription by binding to chromatin.15
Mir-548, a super primate-specific miRNA gene family, has 69 genes located in almost all human chromosomes. As a result of its perfect alignment with human immunodeficiency virus (HIV-1), hepatitis C virus, and hepatitis B virus, miR-548 has become an attractive target for the development of novel antiviral therapeutics, although other studies have demonstrated that mir-548 may play an important role in cancer.16 Both possibilities are intriguing, given that one of the genes in the mir-548 family, mir-548ar, is located on chromosome 11q22.1, which is nearly 70 kb upstream of transcription of NEAT1.
NEAT1 was recently found to be involved in HIV-1 replication,17 while NEAT1 knockdown accompanied a reduction in paraspeckle bodies, suggesting a previously unknown involvement of paraspeckles in regulating the expression of HIV-1 instability element-containing RNAs. Similarly, HIF-2α-dependent transcriptional activation of NEAT1 by induction of nuclear paraspeckle formation that accompanies tumor hypoxia led to cancer cell survival.18 Collectively, these evidences suggest that lncRNA is not only functional, but it may also be a key player in various biological processes that accompany both viral infections and cancer progression.
Fused in sarcoma/translocated in liposarcoma (FUS/TLS) is an RNA-binding protein (RBP) that becomes a primary cause of familial amyotrophic lateral sclerosis (ALS).19 It has been reported that 30 mutations of FUS/TLS attributed to nearly 4% of familial ALS and in rare sporadic patients with no apparent familial history.20 FUS was not only identified as a prominent pathological hallmark in ALS and frontotemporal lobar degeneration,21,22 but it has also been reported to play an important role in many cellular processes, such as alternative splicing,23 embryogenesis,24 and stress response.25 However, little attention has been given to examining FUS in terms of breast development and cancer.
While investigations into the connection between NEAT1 and HIV-1 are ongoing, the underlying mechanisms of NEAT1 in cancer progression and breast tumor cells remain elusive. Our research showed that FUS can bind with NEAT1 physically,26 and we were curious to find that NEAT, which was regulated by miR-548-ar, is required for survival of breast cancer cells and may go on with its function through forming a complex with RBP FUS.
Materials and Methods
Cell culture
Human breast cancer cell lines (MCF-7 and MDA-MB-231) were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin at 37 °C in a humidified atmosphere of 5% carbon dioxide.
Real-time quantitative PCR
Total RNA was extracted from cell lines using TRIzol (Invitrogen), and 2 µg of total RNA was reverse transcribed into first-strand cDNA using the TaKaRa reverse transcription reagent kit according to the manufacturer’s protocol. Quantitative PCR was performed with SYBR Green real-time PCR kit (Toyobo) using the ABI StepOnePlus Real-Time PCR system (Applied Biosystems). All quantifications were performed with GAPDH as the internal standard. The primer sequences were as follows: NEAT1 forward primer 5′CCAGTTTTCCGAGAACCAAA3′, NEAT1 reverse primer 5′ATGCTGATCTGCTGCGTATG3′, FUS forward primer 5′GTGGAGGCAGAGGTGGCATGGGCGG3′, and FUS reverse primer 5′ACATTCTCACCCAGGCCTTGCACAA3′. Results were quantified from three independent experiments.
RNAi
RNAi-mediated knockdown of mRNAs was achieved in all cell types using Stealth RNAi oligos (RiboBio) against NEAT1 (Catalog No. mss205313), FUS (Catalog No. mss208598), or a nonspecific control (Catalog No. 12935-300), at a final concentration of 50 nM. Transfection of RNAi oligos into cell lines was achieved using Lipofectamine 2000 (Invitrogen).
Cell proliferation
siRNA were transfected into all cell lines at a final concentration of 50 nM using Lipofectamine 2000 (Invitrogen). After 24–96 hours, the cells were harvested using trypsin (0.05%) and then manually counted using a Nikon TMS microscope and hemocytometer chamber (Assistent).
Flow cytometry analysis of cell apoptosis
After being treated with siRNA for 48 hours, MCF-7 cells were harvested, suspended in phosphate-buffered saline (PBS), stained with Annexin-V-FITC Apoptosis Detection Kit (BD Biosciences), and analyzed by fluorescence-activated cell sorting analysis, which was carried out using a FACScan flow cytometer (Becton Dickinson) and FlowJo software.
Cell immunofluorescence
In addition to Annexin-V/PI double staining to detect cell apoptosis, activation of apoptosis was also confirmed biochemically by quantification of cleaved caspase-3. Briefly, MCF-7 cells were cultured for 48 hours, treated with siRNA for 12 hours, and then fixed in 4% paraformaldehyde. Cleaved caspase-3 was labeled using a rabbit polyclonal antibody (9661S; Cell Signaling Technology) and Alexa Fluor 488 fluorescent secondary antibody (anti-rabbit IgG (H+L) antibody, KPL). Cell nuclei were stained with DAPI.
Bioinformatic analyses
The likelihood of miRNA binding to NEAT1 was evaluated using the RNAhybrid package.27 After filtering for conservation, five putative target sites were identified. The human FUS/TLS CLIP-seq dataset (SRR556766) was downloaded from European Nucleotide Archive. Barcode and adaptor sequences were removed from reads. Reads were then mapped to the University of California, Santa Cruz human hg19 genome assembly using Bowtie 2.
RNA immunoprecipitation assay
RNA immunoprecipitation assay has been described in our previous study.28 Briefly, 107 MCF-7 cells were grown in 15-cm plates. Cells were harvested in PBS and lysed in 3 mL hypotonic lysis buffer [20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM EDTA, 0.5% NP-40, 0.1% Triton X-100, 1 × EDTA-free protease inhibitor cocktail (ROCHE)] for five minutes on ice. The suspension was then sonicated at 30% amplitude with a microtip in 2-second bursts with 10-second intervals for a total of 30 seconds (Branson Digital Sonifier 250). The lysate was centrifuged at 15,000 × g for 10 minutes at 4 °C. The lysate was incubated for 2 hours at 4 °C with 10 µg of anti-FUS/TLS antibody (A300-293A; Bethyl) precoupled to 50 µL of Protein G Dynabeads (Invitrogen) according to the manufacturer’s instructions. The RNA-protein complexes captured on the beads were washed eight times with 1 mL IsoWB (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% NP-40), then eluted with 200 µL of clear sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 10 mM EDTA and 100 mM DTT) at 25 °C for 5 minutes and subsequently at 95 °C for 2 minutes. The RNA present in the pull-down material was detected by qRT-PCR.
Statistical analyses
All findings are the results of at least three independent experiments. Data are shown as mean ± standard deviation, and the statistical significance of differences between means was assessed by two-tailed t-test. A P-value of 0.05 or less was considered significant.
Results
Knockdown of NEAT1 inhibits growth and induces apoptosis in breast cancer cells
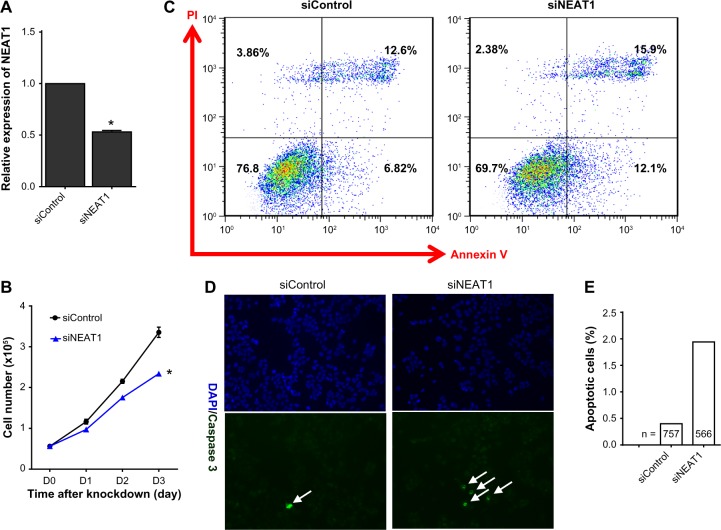
To determine whether NEAT1 promotes cell survival, siRNA was used to knock down NEAT1 expression; relative NEAT1 expression was verified using qRT-PCR (Fig. 1A). Sulforhodamine B assay showed that the knockdown of NEAT1 significantly inhibited cell growth in MCF-7 (Fig. 1B) and MDA-MB231 (Supplementary Fig. 1) cell lines. We noticed that the rate of apoptosis significantly increased after knocking down NEAT1, with the percentage of early apoptotic cells in siNEAT1 cells being ~2-fold greater than siControl. In addition, the percentage of late apoptotic cells increased by more than 25% in siNEAT1 cells (Fig. 1C). These apoptotic effects were further confirmed via immunofluorescence analysis (Fig. 1D); cleaved caspase-3-positive cells were also significantly increased compared with control (Fig. 1E).
Figure 1.
Knockdown of NEAT1 inhibited cell growth and increased cell apoptosis. (A) MCF-7 cells were transfected with NEAT1-specific siRNA (50 nM) and nonspecific control siRNA (50 nM). Knockdown efficiency was determined by qRT-PCR. (B) The time-dependent effect of siRNA on cell growth is shown by the sulforhodamine B assay. Findings are the results of three independent experiments and presented as mean ± SEM. (C) Cell apoptosis was evaluated with Annexin-V and PI double staining at 48 hours by flow cytometry. Values in the lower right quadrant represent the percentage of early apoptotic cells. Values in the upper right quadrant represent the percentage of late apoptotic cells. (D) Immunofluorescence was measured using cleaved caspase-3 (green, apoptotic cells). Nuclei were stained with DAPI (blue). Arrows mark caspase-3-positive cells. (E) The frequency of caspase-3-positive cells is shown. The percent of caspase-3-positive cells was used for chi-square test statistics. Asterisk indicates P< 0.05.
FUS physically interacts with NEAT1
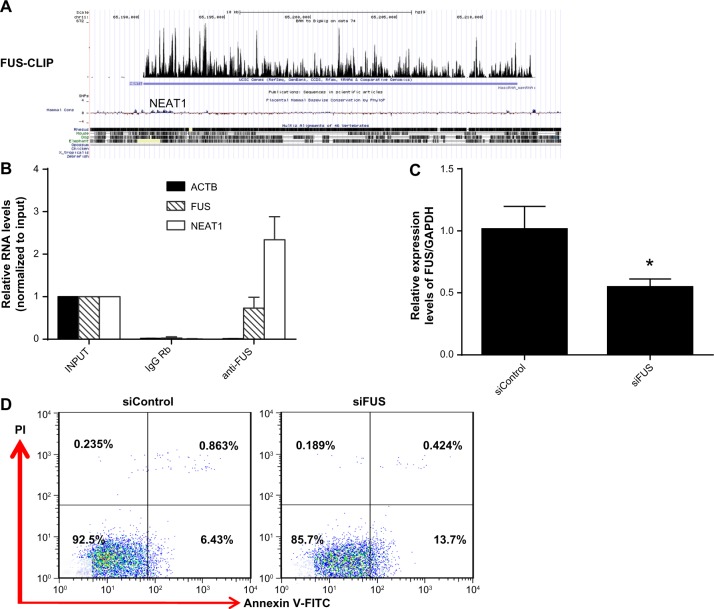
NEAT1 has previously been shown to have significant TDP43 and FUS CLIP (ultraviolet cross-linking and immunoprecipitation) signals,26,29 suggesting that FUS physically binds to NEAT1 (Fig. 2A). We verified this physical interaction by performing RNA immunoprecipitation using a FUS antibody (Fig. 2B). Functionally, administration of si-FUS, whose knockdown efficiencies are shown in Figure 2C, induced cell apoptosis in MCF-7 similar to NEAT1 knockdown (Fig. 2D). These findings suggested that NEAT1 may interact with RBP FUS to influence breast cancer cell apoptosis.
Figure 2.
RBP FUS binds to NEAT1. (A) Short segments of RNA from FUS CLIP-seq were obtained after filtering, processing, and mapping reads to the human genome 19. Wiggle plots of RNA-seq shows the strength of binding with FUS protein. (B) MCF-7 cell lysates were incubated with anti-FUS beads. RNA was then extracted and assessed by qRT-PCR. (C) After transfecting MCF-7 cells with si-FUS, knockdown efficiency was measured by qRT-PCR. (D) Cell apoptosis was analyzed by Annexin-V/PI double staining. ACTB and IgG Rb stand for b-actin and rabbit IgG, respectively.
Overexpressing the miR-548ar downregulates NEAT1 expression
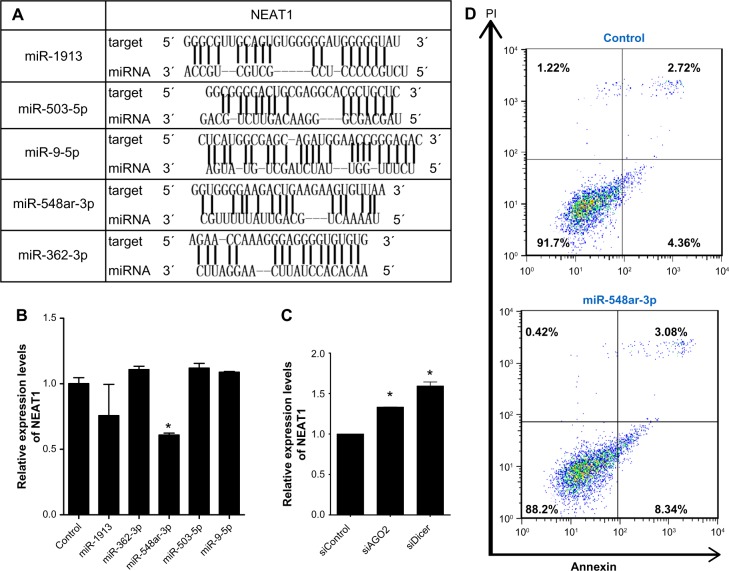
We also further explored that miRNAs could bind to the lncRNA NEAT1 by evaluating the likelihood of mature miRNAs binding to NEAT1 using the RNAhybrid package. After screening based on the scores of conservation, the top five putative miRNAs were identified based on the strength of binding with lncRNA NEAT1 (Fig. 3A). These five miRNA mimics were then synthesized for transfection of MCF-7 cells. Upon overexpressing each candidate miRNA, only miR-548ar demonstrated reduced NEAT1 expression as assessed by qRT-PCR (Fig. 3B). We further found that the expression of NEAT1 was increased after transfecting cells with siRNA of AGO2 or Dicer, which was key to miRNA processing (Fig. 3C). These findings suggest that certain miRNAs can downregulate NEAT1 expression. To verify the regulation of miR-548ar on NEAT1, we transfected cells with the miR-548ar miRNA mimic to determine whether overexpressing miR-548ar also promotes apoptosis. Indeed, our results showed that overexpression of miR-548ar induced cell apoptosis (Fig. 3D). Together, these findings indicate that miR-548ar can downregulate the expression of NEAT1 and induce cell apoptosis.
Figure 3.
miR-548ar interacts with NEAT1. (A) Predicted miRNAs of seed sequences targeting NEAT1 by RNAhybrid. (B) qRT-PCR for NEAT1 RNA levels in MCF-7 cells transiently transfected with synthesized microRNA mimics (20 nM). Findings are the results of three independent experiments and presented as mean ± SEM. (C) Relative expression of NEAT1 after MCF-7 cells were transfected with siAGO2 or siDicer. (D) Apoptosis of MCF-7 cells was detected by FACS staining with Annexin-V 48 hours after transfecting cells with NEAT1 siRNA.
Discussion
Despite the critical roles that lncRNAs may play in gene regulation, the sheer quantity ncRNAs and early stages of the research have made it difficult for researchers to succinctly pinpoint the general working mechanism underlying lncRNAs or their specific interactions. As more lncRNAs are studied and their regulatory activities are investigated, the need to derive a working model for lncRNA becomes more pressing. As we mentioned earlier, there are two major models: the sponge model and the chromatin model. Differing reports have lent support to either model, but our current findings suggest that neither may be fully correct. Alternatively, we propose that our observations of NEAT1 suggest a novel, third model, wherein the RBP FUS and NEAT1 forms a complex, which target on apoptosis signals; the stabilization of this complex then becomes critical in the regulation of chromatin and gene expression.
There are a few fragmented reports dealing with RBPs and cell survival. The RNA-binding motif 5 (RBM5) and 10 (RBM10) were found to promote apoptosis in cancer cells by activating alternative splicing of key death/survival genes.30 Silencing the RBP human antigen R was able to inhibit cell proliferation and increase apoptosis.31 Ectopic expression of RBP poly C-binding protein was found to induce cell cycle arrest in G2 and apoptosis through the cyclin-dependent kinase inhibitor p21.32 Though these lines of evidence do not elucidate the underlying mechanism of RBPs on cancer cell apoptosis, our model suggests that RBPs exert their survival functions by mediating lncRNA and RBP complexes. In practice, this means that lncRNAs can stabilize RBPs or vice versa, a finding that was found during an investigation into liver cancer.33 This model paired with further consistent results may open a new avenue of inquiry for exploring the functions of RBPs.
miRNAs may regulate cell apoptosis in several ways. First, miRNAs can target genes involved in apoptosis. For example, let-7a was shown to regulate the drug-induced apoptosis in cells by targeting caspase-334; miR-21 and miR-15/16 can target the proapoptotic factor B-cell CLL/lymphoma 2 (BCL-2) to inhibit cell apoptosis in glioblastoma and lymphoma.35 Second, miRNAs can target factors that can then influence cell apoptosis. For example, miR-155 directly regulates FOXO3a in the control of breast cancer cell survival, promoting cell death by upregulation of proapoptotic genes, including BCL-2-like 11 (BIM), p27, BCL-2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), and repression of antiapoptotic genes such as FLIP and BCL-XL.36 miR-21 was also reported to negatively regulate F-box protein 11 (FBXO11) in cancer cells, acting as a tumor suppressor and promoting apoptosis by interacting with antiapoptotic gene B-cell CLL/lymphoma 6 (BCL6).37 Although the miR-548 family was not previously reported to be involved in regulating cell apoptosis, we found that miR-548ar could induce cell apoptosis in the breast cancer cell lines MCF-7 and MDA-MB231. Accordingly, we propose that miR-548ar may regulate cell apoptosis by interacting with NEAT1. Although we found that overexpressing miR-548ar downregulated the expression of NEAT1, our luciferase assay demonstrated that miR-548ar could not directly bind to NEAT1 (data not shown). It will be necessary to further characterize the mechanisms of miR-548ar interacting with NEAT1.
In terms of stress, while previous reports demonstrated that NEAT1 deficiency could only induce apoptosis upon hypoxia,18 our data showed that normal oxygen stress disruption of NEAT1 could also serve as an incentive for cell death. This discrepancy may be due to the different strategies used for knocking down expression. For example, our studies employed double-stranded siRNAs that targeted the 5′ end of NEAT1_1, while the antisense oligos in hypoxia stress targeted the 3′ end, raising the possibility that RBP is recognized through the 5′ region. The region of our siRNA-targeted has stronger binding affinity to FUS than the other regions including antisense oligos used in other studies (Supplementary Fig. 2), supporting our hypothesis that the complex of FUS and NEAT1 promotes the survival of breast cancer cells.
Taken together, our research indicated that lncRNA NEAT1 is required for the survival of breast cancer cells. The findings of this study have significant implications regarding our understanding of lncRNA to breast cancer. RBP FUS could physically bind with NEAT1, which may mediate the role of NEAT1 in the survival of breast cancer cells. Besides, NEAT1 could also be regulated by miRNA miR-548ar, which also influences apoptosis in human breast cancer cells. And future work should be focused on the specific mechanism of NEAT1-targeting FUS and miR-548ar to influence cell apoptosis.
Supplementary Materials
Supplementary Figure 1. Knockdown of NEAT1 inhibited cell growth in MDA-MB231. MDA-MB231 cells were transfected with NEAT1-specific siRNA(50nM) and negative control siRNA(50nM). (A) Knockdown efficiency was determined by qRT-PCR. (B) The time-dependent effect of siRNA on cell growth is shown by the SRB assay. Results were quantitated from three independent experiments. Data are presented as means ± SEM.
Supplementary Figure 2. Cell apoptosis could be induced by treated with different si-NEAT1. MCF7 cells were transfected with NEAT1 50nM siRNAs targeted by different position of NEAT1. (A) Knockdown efficiency was determined by qRT-PCR. (B) Cell apoptosis was evaluated with Annexin-V and PI double-staining at 48 h by Flow cytometric analysis. Si-NEAT1–1051 and si-NEAT1–3529 stand for siRNA targeted the 1051 and 3529 position of NEAT1 RNA. Si-NEAT1-GIVEN represents the siRNA which used in the parper of Choudhry H et al, 2014.
Footnotes
ACADEMIC EDITOR: James Willey, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totalled 2276 words, excluding any confidential comments to the academic editor.
FUNDING: This study was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDB13030400) and the National Science Foundation of China (No. 31371502). We thank Andrew Willden of the Kunming Institute of Zoology and Kelly Limoncelli of University of Massachusetts Medical School for assistance in the final manuscript and Kunming Biological Diversity Region Center of Instruments for technical support. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: HK, LZhao, LP, BJ. Analyzed the data: XF, HK, HX. Wrote the first draft of the manuscript: HK, LZhao, BJ. Contributed to the writing of the manuscript: LP. Agree with manuscript results and conclusions: LZou, QY, XS. Jointly developed the structure and arguments for the paper: LP, BJ. Made critical revisions and approved final version: HK, LZhao, BJ. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Wright MW, Bruford EA. Naming ‘junk’: human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5(2):90–8. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeOcesano-Pereira C, Amaral MS, Parreira KS, et al. Long non-coding RNA INXS is a critical mediator of BCL-XS induced apoptosis. Nucleic Acids Res. 2014;42(13):8343–55. doi: 10.1093/nar/gku561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Richards EJ, Zhang G, Li Z-P, et al. Long non-coding RNAs regulated by TGFβ: lncRNA-HIT mediated TGFβ-induced epithelial to mesenchymal transition in mammary epithelia. J Biol Chem. 2015;290(11):6857–67. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kallen AN, Zhou X-B, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–12. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakagawa S, Shimada M, Yanaka K, et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. 2014;141(23):4618–27. doi: 10.1242/dev.110544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gremlich S, Damnon F, Reymondin D, et al. The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development. Placenta. 2014;35(1):44–9. doi: 10.1016/j.placenta.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Standaert L, Adriaens C, Radaelli E, et al. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA. 2014;20(12):1844–9. doi: 10.1261/rna.047332.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura K, Imamachi N, Akizuki G, et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Zeng C, Xu Y, Xu L, et al. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14(1):693. doi: 10.1186/1471-2407-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarty D, Sboner A, Nair SS, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper DR, Carter G, Li P, Patel R, Watson JE, Patel NA. Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARγ2 splicing during adipogenesis in 3T3-L1 Cells. Genes. 2014;5(4):1050–63. doi: 10.3390/genes5041050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose T, Virnicchi G, Tanigawa A, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25(1):169–83. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu B, Ying X, Wang J, et al. Identification of a tumor-suppressive human-specific microRNA within the FHIT tumor-suppressor gene. Cancer Res. 2014;74(8):2283–94. doi: 10.1158/0008-5472.CAN-13-3279. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Q, Chen C-Y, Yedavalli VS, Jeang KT. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4(1):e596–12. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhry H, Albukhari A, Morotti M, et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene. 2014;34(34):4546. doi: 10.1038/onc.2014.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 20.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimoto Y, Nakagawa S, Hirose T, et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain. 2013;6:31. doi: 10.1186/1756-6606-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 23.Polymenidou M, Lagier-Tourenne C, Hutt KR, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14(4):459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sephton CF, Cenik B, Cenik BK, Herz J, Yu G. TDP-43 in central nervous system development and function: clues to TDP-43-associated neurodegeneration. Biol Chem. 2012;393(7):589–94. doi: 10.1515/hsz-2012-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker SJ, Meyerowitz J, James JL, et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int. 2012;60(4):415–24. doi: 10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruger J, Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451–4. doi: 10.1093/nar/gkl243. (2006).10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricci EP, Kucukural A, Cenik C, et al. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol. 2014;21(1):26–35. doi: 10.1038/nsmb.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–8. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson TC, Du L, Janesko-Feldman K, et al. The nuclear splicing factor RNA binding motif 5 promotes caspase activation in human neuronal cells, and increases after traumatic brain injury in mice. J Cereb Blood Flow Metab. 2015;35(4):655–66. doi: 10.1038/jcbfm.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vo DT, Abdelmohsen K, Martindale JL, et al. The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res. 2012;10(1):143–55. doi: 10.1158/1541-7786.MCR-11-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scoumanne A, Cho SJ, Zhang J, Chen X. The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res. 2011;39(1):213–24. doi: 10.1093/nar/gkq778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60(4):1278–90. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 34.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102(39):13944–49. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong W, He L, Coppola M, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285(23):17869–79. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Yang CH, Pfeffer SR, Sims M, et al. The oncogenic microRNA-21 inhibits the tumor suppressive activity of FBXO11 to promote tumorigenesis. J Biol Chem. 2015;290(10):6037–46. doi: 10.1074/jbc.M114.632125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Knockdown of NEAT1 inhibited cell growth in MDA-MB231. MDA-MB231 cells were transfected with NEAT1-specific siRNA(50nM) and negative control siRNA(50nM). (A) Knockdown efficiency was determined by qRT-PCR. (B) The time-dependent effect of siRNA on cell growth is shown by the SRB assay. Results were quantitated from three independent experiments. Data are presented as means ± SEM.
Supplementary Figure 2. Cell apoptosis could be induced by treated with different si-NEAT1. MCF7 cells were transfected with NEAT1 50nM siRNAs targeted by different position of NEAT1. (A) Knockdown efficiency was determined by qRT-PCR. (B) Cell apoptosis was evaluated with Annexin-V and PI double-staining at 48 h by Flow cytometric analysis. Si-NEAT1–1051 and si-NEAT1–3529 stand for siRNA targeted the 1051 and 3529 position of NEAT1 RNA. Si-NEAT1-GIVEN represents the siRNA which used in the parper of Choudhry H et al, 2014.