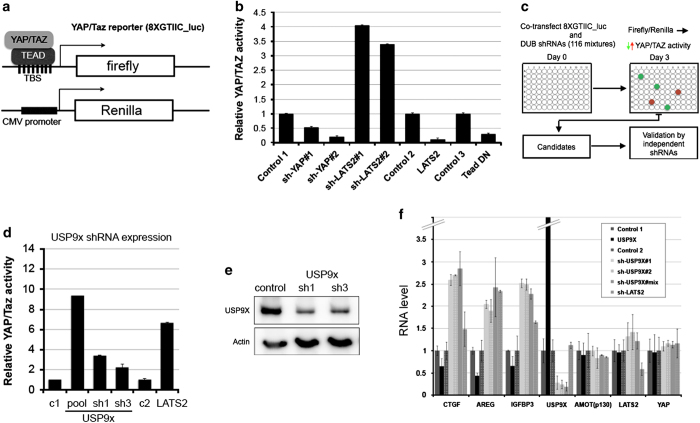
Figure 1.
Identification of USP9x as a regulator of YAP activity. (a) The 8×GTIIC_luc YAP/TAZ reporter contains eight TEAD-binding sites, to control expression of firefly luciferase. CMV-Renilla luciferase provides a control to normalize for transfection efficiency. YAP/TAZ activity was determined by the ratio of firefly/Renilla luciferase after co-transfection of the two plasmids. (b) Luciferase reporter assays showing the effects of changes in Hippo pathway activity. HEK293T cells were transfected to express the luciferase reporters together with shRNA vectors to deplete YAP, LATS2 or with control shRNAs. Cells were also transfected to overexpress LATS2 or a dominant negative form of TEAD vs appropriate empty vectors as controls. Data represent the mean of three independent transfection experiments±s.d. (c) Summary of the RNAi screen workflow. (d) Luciferase reporter assays showing the effects of changes in USP9x activity. HEK293T cells were transfected to express the firefly and luciferase reporters together with an shRNA pool and two individual shRNAs targeting USP9x. Data represent the mean of three independent replicates±s.d. (e) Immunoblot showing the efficacy of shRNA-mediated depletion of USP9x protein. Upper panel probed with anti-USP9x. Lower panel probed with anti-Actin to control for loading. (f) Quantitative PCR was used to measure YAP target transcript levels in HEK293T cells transfected to express the indicated shRNAs. USP9x was depletion using the shRNA pool and two individual shRNAs. shRNA-mediated depletion of LATS2 was used as a control for the effect of increasing YAP/TAZ activity. RNA levels for UAP9x and LATS2 are shown to monitor shRNA efficiency. To test the effect of USP9x overexpression, HEK293T cells were transfected to express V5-tagged USP9x or with an empty vector control. RNA was harvested 36 h after transfection. GAPDH was used for normalization. YAP target levels increased, while YAP mRNA was unchanged. Data represent the average of three independent experiments±s.d.