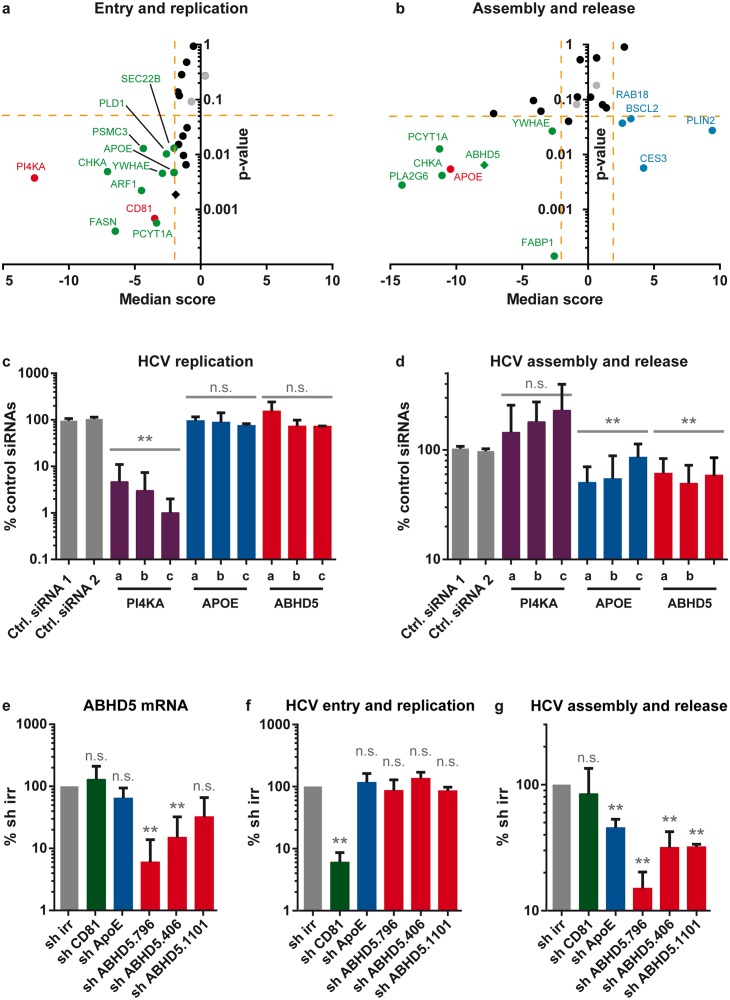
Fig 1. Identification of ABHD5 as a new host factor for HCV production.
(a, b) A rational siRNA screen was designed to identify host factors involved in the lipid metabolism and participating in the HCV replication cycle (see S2 Fig) with readouts for HCV entry and replication (a) or assembly and release (b). For each graph, the p-value is plotted against the median score. A maximal p-value of 0.05 together with a mean score superior to 2 (blue dots, antiviral factors) or inferior to -2 (green dots, proviral factors) was considered highly significant. CD81, PI4KA and APOE controls are shown in red in the relevant graph and the non-targeting negative control siRNAs in grey. ABHD5 is depicted with a diamond. Yellow dotted lines indicate our statistical thresholds. (c, d) ABHD5-specific siRNAs used in the initial screen as a pool (panels a and b) were transfected individually into HCV RNA-transfected cells. Their specific effect on HCV RNA replication (panel c, corrected for cell viability effects) and progeny virion production (panel d, corrected for HCV RNA replication effects) is depicted after normalisation to the average value of two non-targeting siRNAs. Note that statistics were performed at the gene level. (e- g) Effect of ABHD5-specific shRNAs on ABHD5 gene expression (e), HCV entry and replication (f) and HCV assembly and release (g). (e) ABHD5 mRNA levels were quantified by qRT-PCR at the time of virus harvest. (f) HCV entry and replication were determined by the RLuc activity in the producer cell lysates at the same time point and corrected for the effects on cell viability. (g) The efficiency of HCV production was evaluated by the RLuc activity in target cells infected with the supernatant of shRNA-transduced and JcR-2a-infected producer cells, and corrected for the shRNA effects on HCV entry and replication.