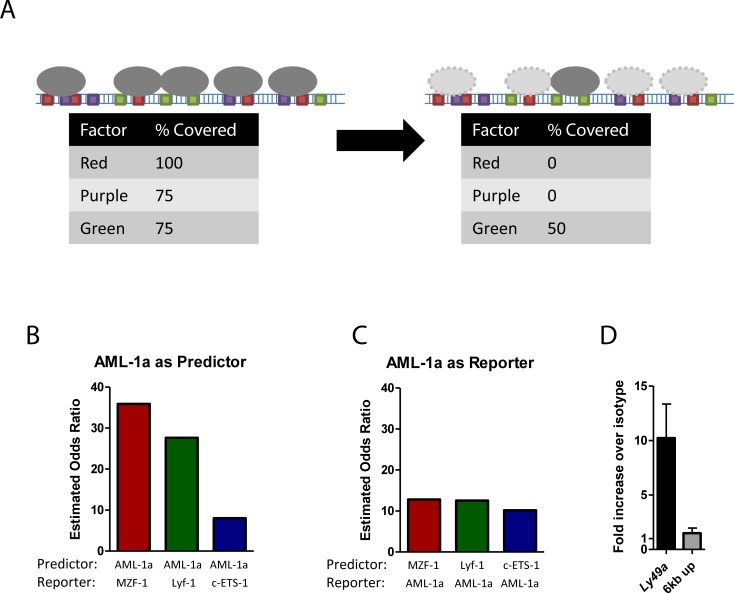
Fig 5. Confounding variables unlikely to account for AML-1a-specific nucleosome depletion in Ly49A.
(A) Schematic highlighting the possibility of confounding variables impacting transcription factor/nucleosome relationship studies. If an event occurs to deplete all nucleosomes covering a red factor, the coverage of the red factor drops from 100% to 0%. However, the coverage of all purple factors drops from 75% to 0%, and green from 75% to 50%. Taking nucleosome coverage as the readout, it is impossible to determine whether the red, purple, or green factors ‘caused’ the nucleosome depletion. However, if red depletions are always correlated with purple depletions, while purple depletions only occasionally predict red depletions, it is likely that red is causing the nucleosome remodeling. (B-C) Logistic regression analysis was performed for each of the four most depleted transcription factors from 4C (AML-1a, c-ETS-1, Lyf-1, and MZF-1), using (B) AML-1a as the predictor, or (C) AML-1a as the reporter. In each case, AML-1a was a better predictor of the other two nucleosome-preferring factors, Lyf-1 and MZF-1, than either were of AML-1a. AML-1a performed equally well as predictor or reporter against c-ETS-1, a factor that had no preference for nucleosome coverage, and so was unlikely to be involved as a confounding variable. (D) Chromatin immunoprecipitation (ChIP) using anti-AML-1 or an irrelevant isotype control was performed on RMA chromatin extract. ChIP using the anti-AML-1 antibody resulted in about a 10-fold enrichment of the Ly49A promoter region when compared to ChIP using the isotype control, indicating that AML-1 is likely bound to the promoter. As a control, a region of DNA approximately 6 kbp upstream of the Ly49A promoter displayed no such enrichment.