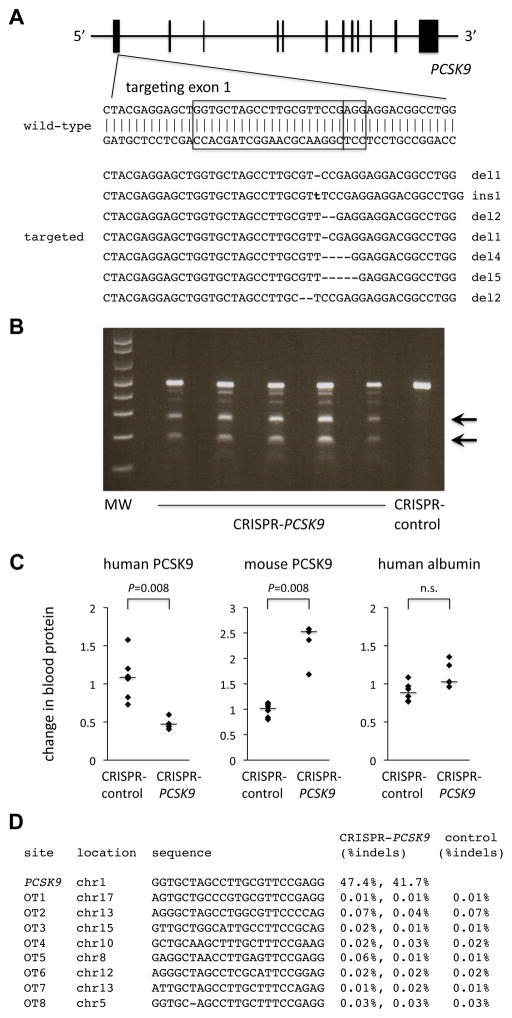
Figure 1.
CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo. A, Targeting of a sequence in exon 1 of the human PCSK9 gene. The boxes indicate the 20-bp sequence matching the protospacer and the 3-bp protospacer-adjacent motif (PAM). The seven targeted sequences shown below the wild-type sequence reflect the seven most common mutations (in descending order of frequency) detected by deep sequencing of the locus in human hepatocytes targeted in vivo in chimeric liver-humanized mice (see Supplemental Dataset for more information). B, Surveyor assays performed with genomic DNA from liver samples taken from mice 4 days after receiving an adenovirus expressing Cas9 and the PCSK9 guide RNA (CRISPR-PCSK9) or a control adenovirus (CRISPR-control). Arrows show the cleavage products resulting from the Surveyor assays; the intensity of the cleavage product bands relative to the uncleaved product band corresponds to the mutagenesis rate. C, Relative changes in blood human PCSK9 protein levels, blood mouse PCSK9 protein levels, and blood human albumin levels (post-treatment divided by pre-treatment levels) in chimeric liver-humanized mice receiving CRISPR-control virus (n = 6 mice) or CRISPR-PCSK9 virus (n = 5 mice). The bars indicate the median values for the relative changes within the groups. The Mann–Whitney U test was performed to compare the relative changes in the two groups. D, Indel rates at on-target and off-target sites from next-generation DNA sequencing of liver samples from post-treatment mice.