Table 1. Structures of the MK2 kinase and its complexes stored in the PDB.
| PDB entry Release date |
Chain | Solved sequence/ Protein Constructa |
Resolutionb | Ligand | Binding modec | Reference |
|---|---|---|---|---|---|---|
| 1kwp 2002-09-18 |
A, B | 46-385 47-400 |
2.80 Å | apoenzyme | 36 | |
| no PDB entry 2003 |
45-371 | 3.0 Å |
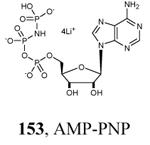
|
|
37 | |
| 1ny3 2003-10-14 |
A | 46-345 41-364 |
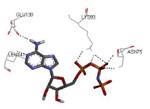
3.00 Å | ADP | see Figure 2 | 35 |
| 1nxk 2003-10-14 |
A, B, C, D | 42-345 41-364 |
2.70 Å |
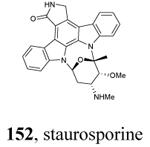
|
|
35 |
| 2okr 2007-02-06 |
C, F (MK2) A, D (p38α) |
370-393 370-400 |
2.0 Å | p38α-MK2 heterodimer | 31 | |
| 2onl 2007-02-06 |
C, D (MK2) A, B (p38α) |
46-393 Full length |
4.0 Å | p38α-MK2 heterodimer | 31 | |
| 2jbod 2007-03-20 |
A | 44-347 41-364 |
3.10 Å |
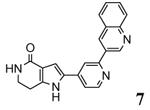
|
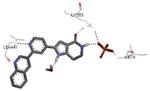
|
63 |
| 2jbpe 2007-03-20 |
A, B, C, D, E, F, G, H, I, J, K, L |
46-350 41-364 |
3.31 Å |
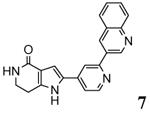
|
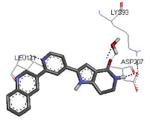
|
63 |
| 2oza 2007-04-03 |
A (MK2) B (p38α) |
51-390 47-400 |
2.70 Å | p38α-MK2 heterodimer | 30 | |
| 2p3g 2007-06-12 |
X | 45-364 45-371 |
3.80 Å |
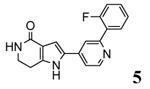
|
|
46 |
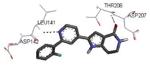
| 2pzy 2007-07-31 |
A, B, C, D | 42-346 41-364 |
2.90 Å |
|
|
60 |
| 3fpm 2009-04-07 |
A | 44-345 41-364 |
3.30 Å |
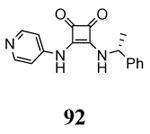
|
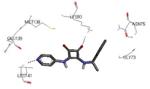
|
77 |
| 3fyk 2009-04-07 |
X | 45-364 45-371 |
3.50 Å |
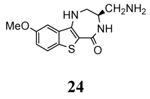
|
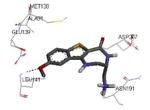
|
51 |
| 3fyj 2009-04-07 |
X | 45-364 45-371 |
3.80 Å |
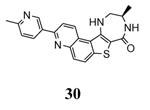
|
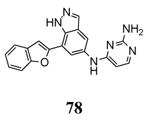
|
52 |
| 3kc3 2010-01-12 |
A, B, C, D, E, F, G, H, I, J, K, L |
43-347 41-364 |
2.90 Å |
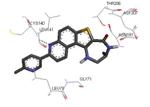
|
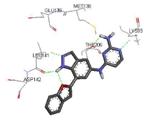
|
42 |
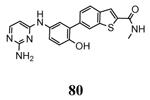
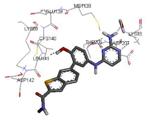
| 3ka0 2010-01-12 |
A | 47-364 47-366f |
2.90 Å |
|
|
42 |
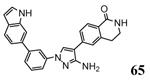
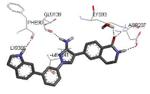
| 3kga 2010-01-26 |
A | 47-364 47-364 |
2.55 Å |
|
|
43 |
| 3gok 2010-03-02 |
A, B, C, D, E, F, G, H, I, J, K, L |
46-350 47-400 |
3.20 Å |
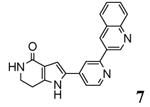
|
unpublished resultsg |
|
| 3a2c 2010-05-12 |
A, B, C, D, E, F, G, H, I, J, K, L |
47-344 41-364 |
2.90 Å |
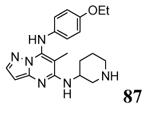
|
|
38 |
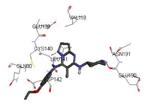
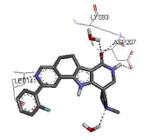
| 3m2w 2010-07-28 |
A | 47-364 47-364 |
2.41 Å |
|
|
73 |
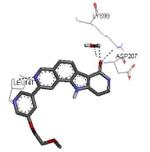
| 3m42 2011-03-23 |
A | 47-364 47-364 |
2.68 Å |
|
|
72 |
| 3r2y 2011-05-25 |
A | 46-365 41-364 |
3.00 Å |
|
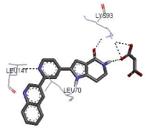
|
64 |
| 3r30 2011-05-25 |
A | 46-365 41-364 |
3.20 Å |
|
|
64 |
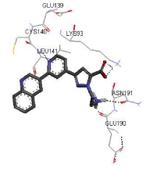
| 3r2b 2011-05-25 |
A, B, C, D, E, F, G, H, I, J, K, L |
47-345 41-364 |
2.90 Å |
|
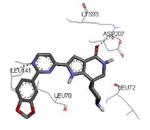
|
64 |
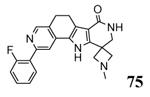
| 3wi6 2013-12-18 |
A, B, C, D, E, F |
46-347 41-364 |
3.00 Å |
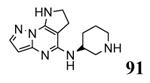
|
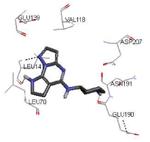
|
40 |
| 4tyh 2015-07-22 |
A (MK2) B (p38α) |
51-400 | 3.00 Å | p38-MK2 heterodimer with a p38 inhibitor |
101 |
When more than one MK2 chain has been crystallized, the solved sequence and protein construct are referred to the first chain listed in the previous column. Solved sequence: the amino acid sequence listed in the PDB structure. Protein construct: the MK2 protein construct used in the study.
All the structures have been obtained by X-ray diffraction.
Graphical representations of the binding mode of MK2 inhibitors have been elaborated with Discovery Studio 3.0 Visualizer software (Accelrys Software, Inc.) by the corresponding PDB files. The coordinates of the complex between MK2 and AMP-PNP have been taken from the patent text (https://www.google.com.ar/patents/EP1578687A2?cl=en, accessed August 7, 2015), edited to correct typos probably due to a previous scanning from original patent documents, and then converted to a PDB file by means of an in house script.
Bipyramidal crystals, by soaking.
Orthorhombic crystals, by co-crystallyzation
T222E mutant.
Scheich, C.; Smith, M. A.; Barker, J. D.; Kahmann, J.; Hesterkamp, T.; Schade, M. Unpublished results.
