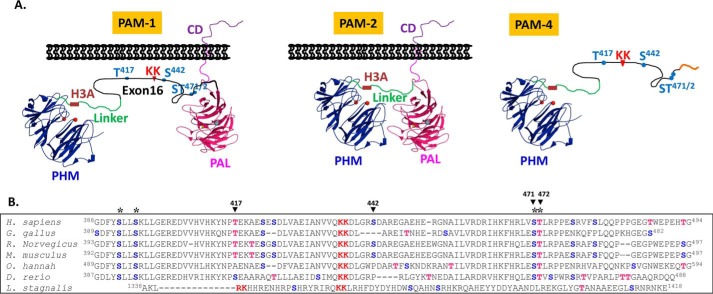
FIGURE 1.
Vertebrate PAM exon 16 has well conserved potential O-glycosylation sites. A, two major PAM isoforms in rodents and human are shown, rodent and human PAM-1 include exon 16 (105 and 107 amino acids, respectively; black line). The protease-resistant PHMcc (blue) is followed by a protease-sensitive 36-amino acid linker (green) that includes a pH-sensitive His cluster (H3A). In PAM-4, a minor splice variant, transcripts terminate in the intron following exon 16 (1). The PALcc (red) is shown, along with the cleavage site in exon 16 (KK) and the Ser/Thr sites explored by mutagenesis. B, ClustalW alignment of the alternatively spliced exon from several vertebrate PAM proteins; the conserved PC cleavage site (KK, in red) and conserved potential O-glycosylation sites (*) are shown: Ser (blue); Thr (red). Accession numbers for the sequences shown: Homo sapiens, NP_000910.2; Gallus gallus, XP_424857.3; Rattus norvegicus, NP_037132.2; Mus musculus, NP_038654.2; Ophiophagus hannah, ETE72045.1; Danio rerio, XP_699436.4; Lymnaea stagnalis, AAD42258.1. Although L. stagnalis PAM included an alternatively spliced exon with a prohormone convertase cleavage site, potential O-glycosylation sites were not conserved.