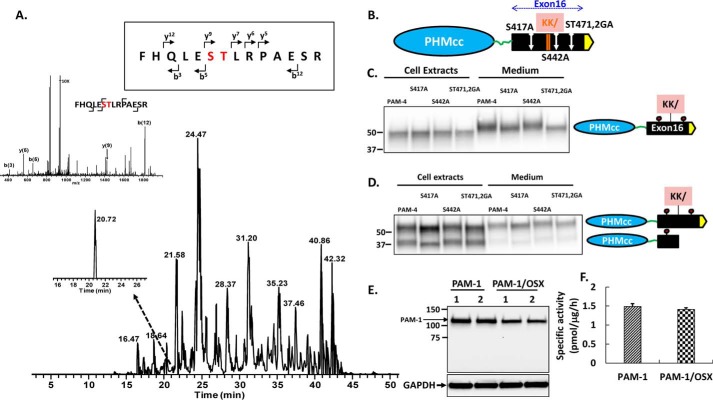
FIGURE 2.
Identification of O-glycosylation sites in exon 16. A, AtT-20 PAM-4 (52 kDa) was subjected to in-gel trypsin digestion. The MS/MS fragment spectrum, along with fragment assignments for O-glycosylated rPAM(466–479), are shown; brackets indicate the b and y ion assignments. Both the y(9) and b(12) fragment ions with the GalNAc modification were detected. B, diagram showing the location of each site-directed mutation relative to the cleavage site in exon 16. PAM-4 and mutant PAM-4 were expressed transiently in pEAK and AtT-20 cells. Using an antibody to the exon 16 region, the apparent molecular weights of the PAM-4 proteins in the media and in cell lysates were compared. For pEAK cells (C), a decrease in the mass of PAM-4/S417A and PAM-4/S471G/T472A versus PAM-4 was apparent in the medium. For AtT-20 cells (D), a decrease in the mass of PAM-4/S417A and S471G/T472A (ST471,2GA) was apparent in intact PAM-4 in both cells and media. The decreased mass of PAM-4/S417A was also apparent in the cleaved protein, which no longer contains S471G/T472A. E, PAM-1 and PAM-1/OSX were transiently expressed in duplicate wells of pEAK cells; lysates (5 μg of protein) were fractionated by SDS-PAGE and visualized using antibody to PHM. F, specific activity of PHM in lysates prepared from pEAK cells transiently expressing PAM-1 or PAM-1/OSX (n = 3).