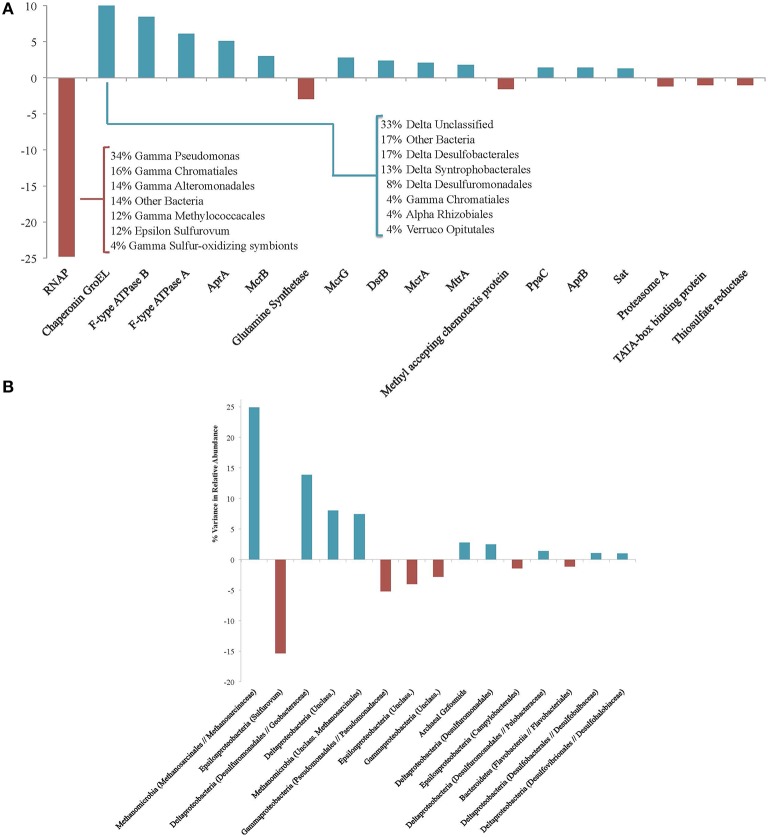
Figure 3.
(A) Protein functional groups accounting for at least 1% of variance between the enriched and unenriched #5133 15N T160d protein pools. Positive values reveal protein types that are more abundant in the enriched fraction; negative values indicate those more prevalent in the unenriched fraction. Inset table shows the phylogenetic affiliations of unenriched RNAP and enriched GroEL proteins. (B) Phylogenetic associations accounting for at least 1% of variance between the enriched and unenriched #5133 15N T160d protein pools. Positive values reveal phylogenetic assignments whose protein products are more abundant in the enriched fraction; negative values indicate those more prevalent in the unenriched fraction. Phylogenetic assignments were made at the family level; higher level assignments are provided if no family-level specificity was available, and genus-level assignments are provided if no other genera were observed in the same family. Archaeal Gzfosmids are ANME-affiliated sequences reported by Hallam et al. (2004).