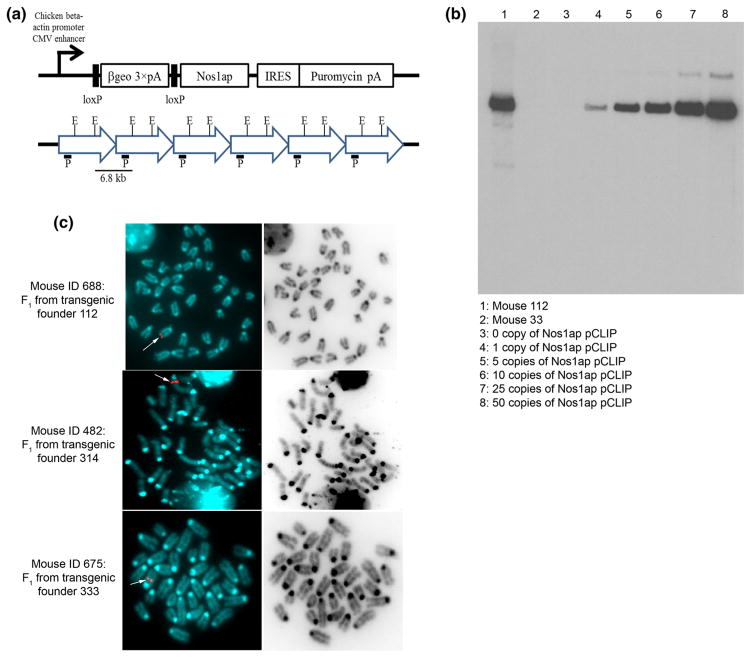
Fig. 1.
Generation of cre recombinase-conditional Nos1ap over-expression transgenic mice. a Nos1ap pCLIP transgene construct (adapted from George et al. 2007), and representation for multiple copy head-to-tail insertion of the transgene and the expected 6.8 kb Southern band with lacZ probe (P) and EcoRV (E) digestion; βgeo β-galactosidase-neomycin fusion gene, pA signal, IRES internal ribosome entry site; b identification of transgenic founder by Southern blotting. Southern blotting performed on mouse tail genomic DNA digested with EcoRV and probed using lacZ specific probe. Lanes 3–8 have wild type mouse tail genomic DNA spiked with zero or multiple copies, as indicated, of Nos1ap pCLIP plasmid DNA per diploid genome. Mouse 112 was selected as a founder line for further experiments; c single site transgene integration in cre recombinase-conditional Nos1ap over-expression transgenic lines. Metaphase FISH using Nos1ap pCLIP plasmid specific probe performed in cultured peripheral blood cells of F1 mice derived from cross between 112, 314, and 333 transgenic founders and FVB mice. Red dots in the left panel (white arrow) indicate site of integration on chromosome 4 (top), chromosome 12 (middle) and chromosome 2 (bottom) for 112, 314 and 333 lines, respectively