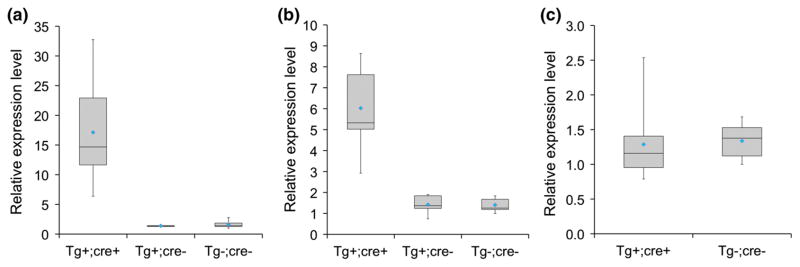
Fig. 2.
Nos1apTg; Mlc2v-cre (Tg+;cre+) mice from two transgenic lines overexpress Nos1ap transcript in left ventricles. a Box-and-Whisker plots showing relative expression levels for Nos1ap transcript in left ventricles from Tg+;cre+ (n = 10), Tg+;cre− (conditional transgene only, n = 3) and wild type control littermates (Tg−;cre−, n = 10) mice derived from transgenic founder 112. Expression of Nos1ap transcript was higher in Tg+;cre+ mice as compared to Tg−;cre− mice, t(18) = 5.77, p <0.001 and expression of Nos1ap transcript was not significantly different between Tg+;cre− and Tg−;cre−mice, t(11) = 0.66, p = 0.52; b same as a, mice derived from transgenic founder 333 (Tg+;cre+ n = 7, Tg+;cre− n = 5, Tg−;cre− n = 7). Expression of Nos1ap transcript was higher in Tg+;cre+ mice as compared to Tg−;cre− mice, t(12) = 5.98, p <0.001 and expression of Nos1ap transcript was not significantly different between Tg+;cre− and Tg−;cre− mice, t(10) = 0.07, p = 0.94; c Same as a, mice derived from transgenic founder 314 (Tg+;cre+ n = 8, Tg−;cre− n = 7). No significant difference in expression level was observed between the two groups of mice derived from transgenic founder 314, t(13) = 0.22, p = 0.83