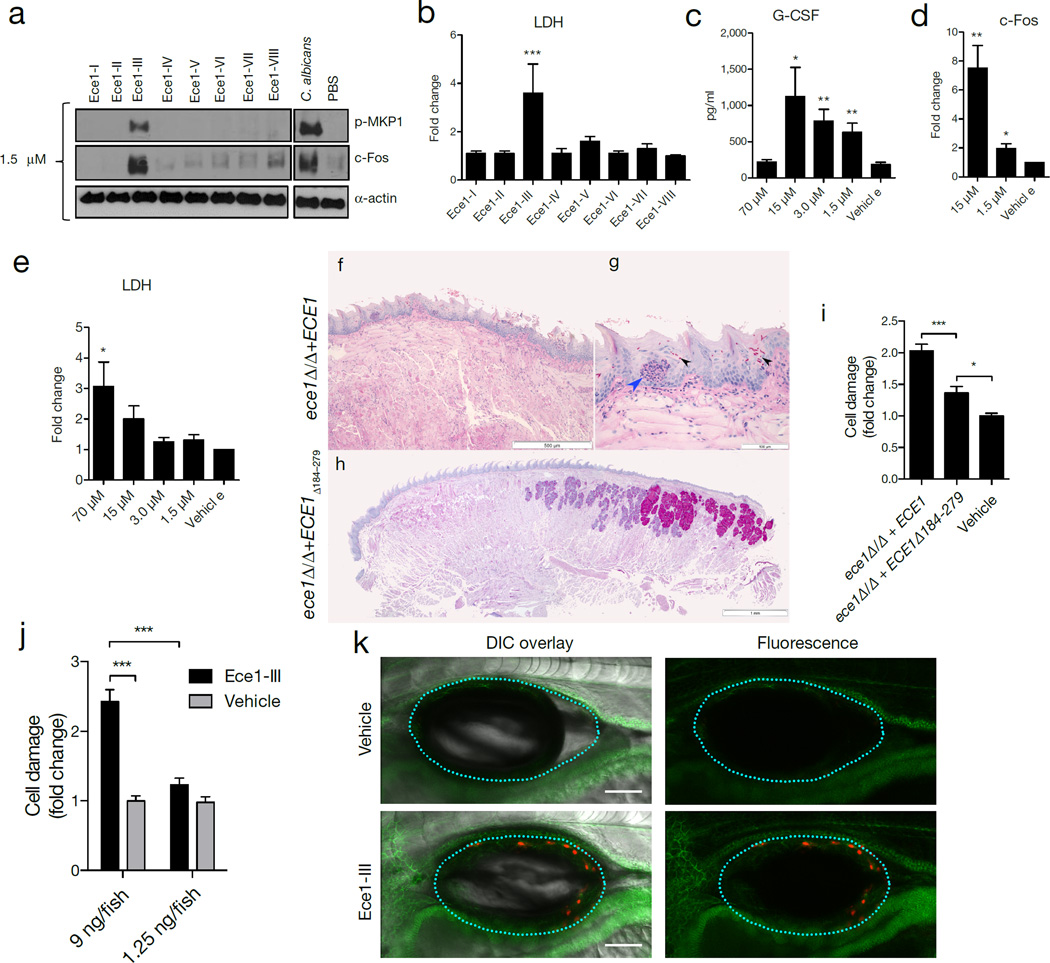
Figure 2. Ece1-III62–93 is the active region of Ece1p and is required for TR146 cell activation and mucosal C. albicans infection.
(a) Induction of p-MKP-1 and c-Fos 2 h post-stimulation (p.s.) with Ece1 peptides at 1.5 µM. (b) LDH release 24 h p.s. with 70 µM Ece1 peptides. (c) Induction of G-CSF 24 h p.s. with Ece1-III62–93 (d) c-Fos DNA binding induction 3 h p.s. with sub-lytic concentrations of Ece1-III62–93 (e) LDH release 24 h p.s. with Ece1-III62–93 (f-h) PAS stained tongue sections from mice subjected to OPC, 2 d p.i. with (f, g) C. albicans ece1Δ/Δ+ECE1 (x25 and x200) or (h) ece1Δ/Δ+ECE1Δ184–279. Invading hyphae (black arrows) and infiltrating inflammatory cells (blue arrow) are shown. (i) Damaged cells in a zebrafish swimbladder 24 h p.i. with C. albicans ece1Δ/Δ+ECE1 (n (number of fish) = 44), ece1Δ/Δ+ECE1Δ184–279 (n = 58) or vehicle (n = 58). (j) Damaged cells in zebrafish swimbladders after stimulation with 9 ng (n = 51) or 1.25 ng (n = 56) Ece1-III62–93, or vehicle (40% DMSO, n = 54 and 5% DMSO, n = 55). (k) Co-localization of adherens junctions (α-catenin-citrine) with Ece1-III62–93-damaged cells (Sytox Orange-positive cells) in a zebrafish swimbladder. Data are representative (a, f-h, k) or mean (b-e, i-j) of three biological replicates (a-d) or ten mice or fish (f-h, k). Error bars show ± SEM. Data were analyzed by one-way ANOVA (b, c, e) paired T test (d) or Kruskal-Wallis (i, j). * = P < 0.05, ** = P < 0.01, *** = P < 0.001 (compared with vehicle control unless otherwise indicated). For gel source data, see Supplementary Figure 1.