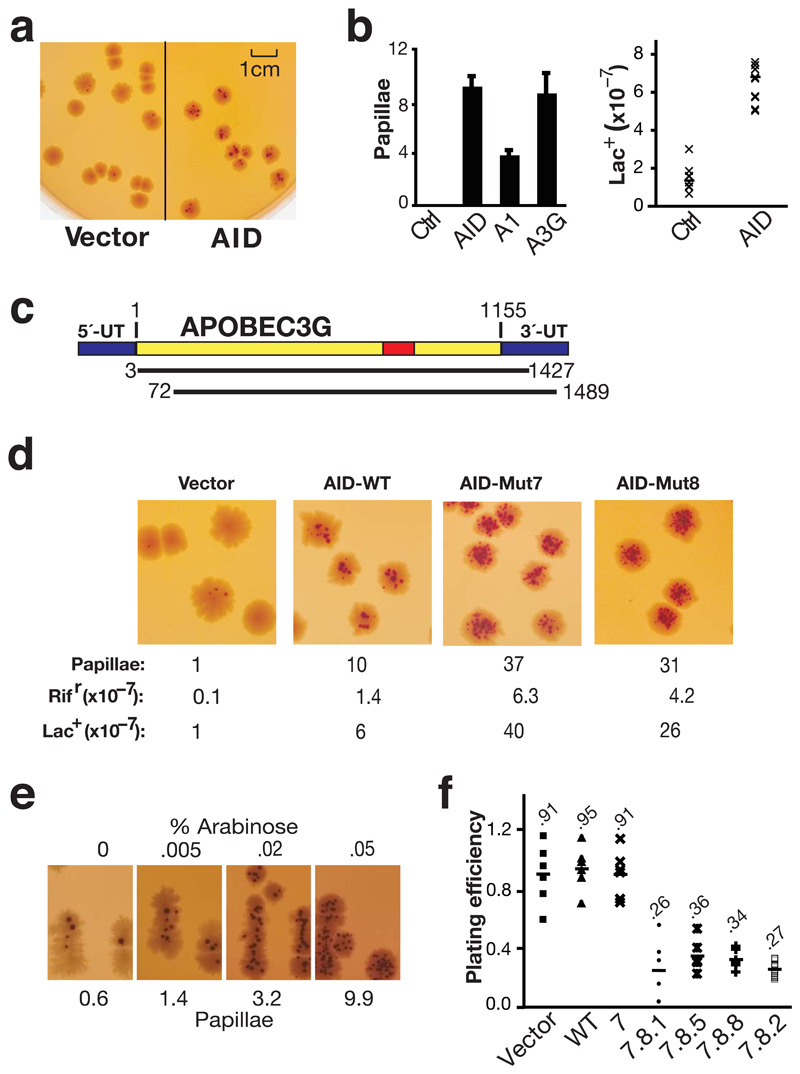
Figure 1.
Papillation screen for active mutators. (a) Empty vector or AID-transformed CC102 was plated on MacConkey-lactose agar and grown at 37 °C for 4 days. (b) Frequency of reversion to Lac+ as determined by the average number papillae per colony on MacConkey-lactose agar (left panel; histogram showing the mean and s.d. of three experiments)) or by growth on minimal lactose plates (right panel: fluctuation analysis on six clones each) in CC102 transformants expressing vector control (Ctrl), AID, APOBEC1 (A1) or APOBEC3G (A3G). (c) Depiction of the two APOBEC3G cDNAs obtained by screening a human spleen cDNA library (50,000 colonies) for papillation on CC102. The wild type full length APOBEC3G mRNA is shown at the top and the structures of the two cDNAs below. The red shows the catalytic active site. The blue shows the untranslated region (UT). Nucleotide residues are numbered relative to the start of the open reading frame (+1). (d) Comparison of papillation (and mutation frequencies on lac and rifampicin) by wild type AID and by upmutants Mut7 (K10E/E156G) and 8 (K34E/K160E). (e) Papillation by AID Mut1.1 expressed from plasmid pBAD30 as a function of the concentration (% w/v) of L-arabinose (inducer). (f) Bacterial titers in cultures grown to saturation in LB/Amp under conditions of IPTG induction relative to the titers obtained from cultures grown in the absence of induction is presented for CC102 transformants expressing different AID upmutants.