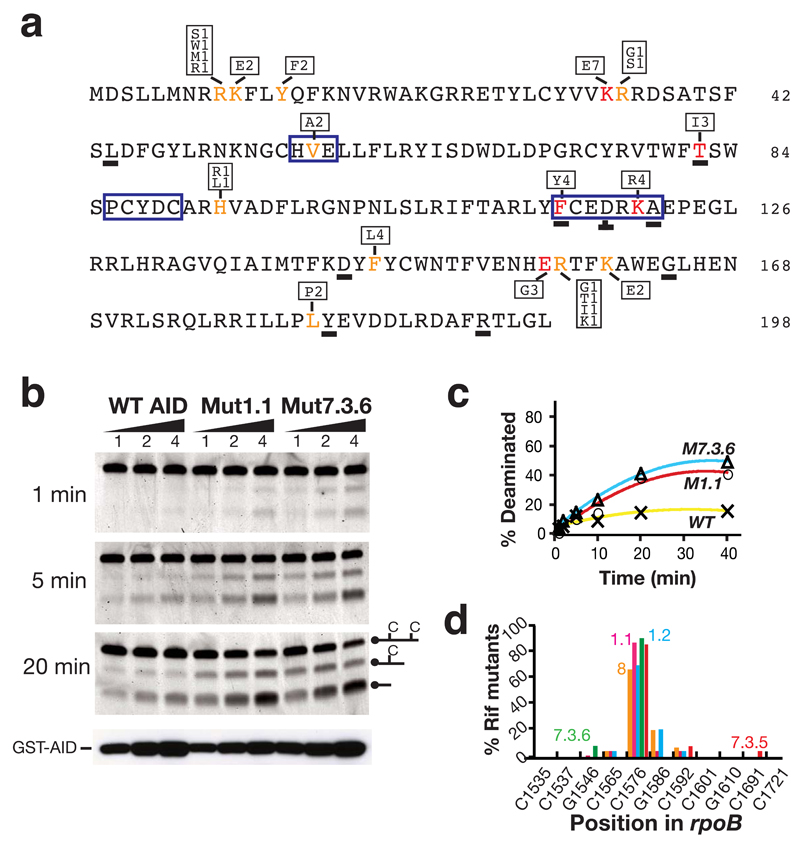
Figure 3.
Nature of the AID upmutations. (a) Primary sequence of human AID with upmutations identified in color. Mutations at residues in red constitute the sole difference between at least one pair of AID sequences resulting in > 2-fold increase in mutation frequencies at rpoB. Mutations at residues in orange have been identified in multiple independent upmutants but in the presence of other substitutions. The box above or below the highlighted residues shows the identity of the substitution mutations and the frequency with which each substitution was detected in the total of nine independent libraries. Underlined letters identify residues where the corresponding position in fugu AID also appears to be a site of selected upmutation, as judged by the fact that it is either the sole mutation or in multiple fugu upmutants. The zinc-coordination motifs (HVE, PCYDC) and regions of suggested polynucleotide contact (FCEDRKA)11–13, are highlighted by a blue box. (b) Deaminase activity of semi-purified GST-AID fusion proteins (100, 200, 400 ng) was analysed on an oligonucleotide substrate at indicated time points. Protein abundance was monitored by Western blot analysis using anti-AID antibody. (c) Quantification of extent of deamination over time. (d) The target specificity of the upmutants as judged by the spectrum of rpoB mutations in Rifr colonies. Transition mutations at any one of 11 C:G pairs within rpoB can give rise to Rifr: the distribution of such mutations amongst Rifr colonies is shown for AID upmutants Mut8 (orange), 1.1 (pink), 1.2 (blue), 7.3.5 (red) and 7.3.6 (green).