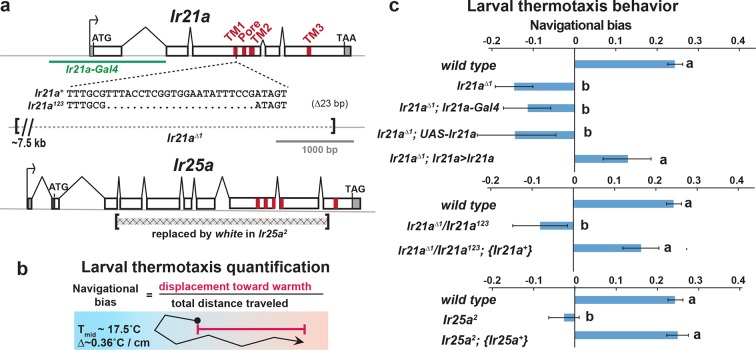
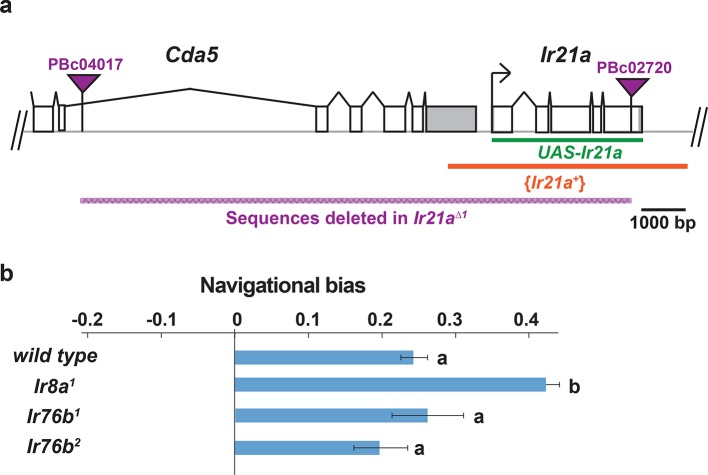
Figure 2. Larval cool avoidance requires Ir21a and Ir25a.
(a) Sequence alterations in Ir21a and Ir25a alleles. Ir21a regulatory sequences present in Ir21a-Gal4 are denoted in green and regions encoding transmembrane domains (TMs) and pore region in red. Additional details provided in Figure 2—figure supplement 1. (b) Thermotaxis is quantified as navigational bias. Cool avoidance behavior was assessed by tracking larval trajectories on a ~0.36˚C/cm gradient extending from ~13.5˚C to ~21.5˚C, with a midpoint of ~17.5˚C. (c) Cool avoidance requires Ir21a and Ir25a. Ir21a>Ir21a denotes a wild type Ir21a transcript expressed under Ir21a-Gal4 control. {Ir21a+} and {Ir25a+} denote wild type genomic rescue transgenes. Letters denote statistically distinct categories (alpha=0.05; Tukey HSD). wild type, n=836 animals. Ir21a∆1, n=74. Ir21a∆1;Ir21a-Gal4, n=48. Ir21a∆1;UAS-Ir21a, n=10. Ir21a∆1;Ir21a>Ir21a, n= 88. Ir21a∆1/ Ir21a123, n=71; Ir21a∆1/ Ir21a123; {Ir21a+} n=70; Ir25a2, n =100. Ir25a2; {Ir25a+} n= 247. Additional mutant analyses provided in Figure 2—figure supplement 1.