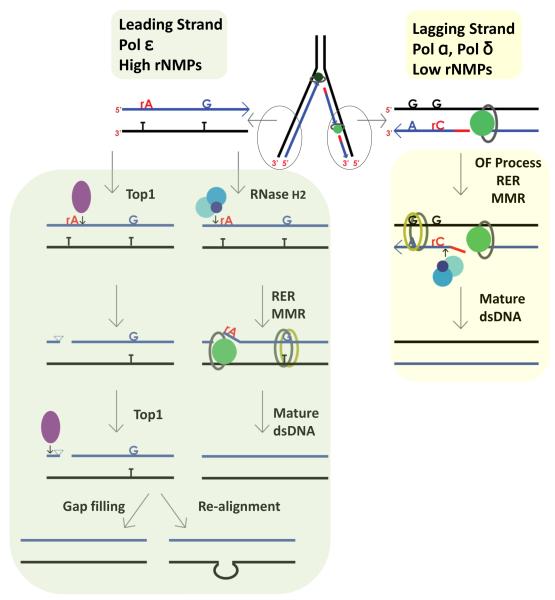
Figure 3. Key Figure. Processing of rNMPs in leading and lagging strand.
Leading strand replicase Pol ε (dark green circle associated with PCNA represented as grey oval) incorporates high number of rNMPs that are used as strand discrimination for newly synthesized DNA. In yeast Top1 can cleave rNMPs, although in a minor pathway that is only evident in the absence of RNase H2. Top1 (represented as purple filled oval) cleaves at the 3′-end of the rNMP forming a 2′-3′ cyclic phosphate that creates unligatable ends. A second Top1 cleaving a few nucleotides upstream would generate a gap that could be filled by the replicative polymerase in an error-free manner. In a sequence with a tandem repeat Top1 could induce realignment in a mutagenic way. RNase H2 (represented as three blue circles) cleaves at the 5′-end of the rNMP (red rA) initiating the RER process that includes strand displacement synthesis (represented as green circle associated with PCNA represented as grey oval), flap removal and ligation identical to OF maturation. The nick created in the DNA by RNase H2 at the rNMP can be used by the MMR system (represented as green and yellow ovals) to access mispaired nucleotides in a strand-specific manner. Lagging strand replicase Pol δ (represented as green circle associated with PCNA represented as grey oval) incorporates low number of rNMPs. The discontinuous nature of lagging strand DNA synthesis permits the loading of the MMR system before OF maturation. RER initiated by RNase H2 can occur concomitant with OF processing or independently. Black represents the template strand, blue the newly synthesized DNA and red the RNA primer of OF.