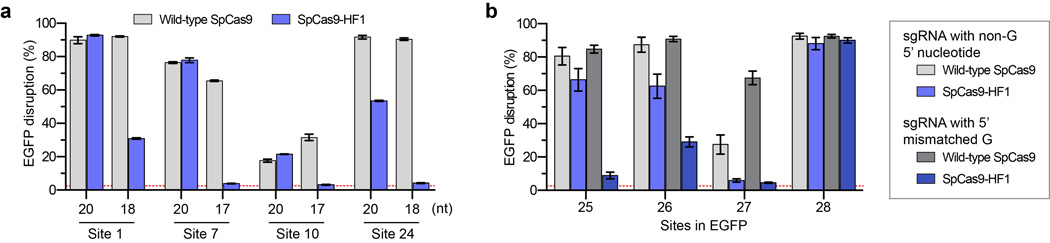
Extended Data Figure 7. Activities of wild-type SpCas9 and SpCas9-HF1 with truncated and 5’ mismatched sgRNAs14.
a, EGFP disruption activities of wild-type SpCas9 and SpCas9-HF1 using full-length or truncated sgRNAs. b, EGFP disruption activities of wild-type SpCas9 and SpCas9-HF1 using sgRNAs that encode a matched 5’ non-G nucleotide or an intentionally mismatched 5’ G nucleotide. For both panels, error bars represent s.e.m. for n = 3, and the mean level of background EGFP loss observed in control experiments is represented by the red dashed line.