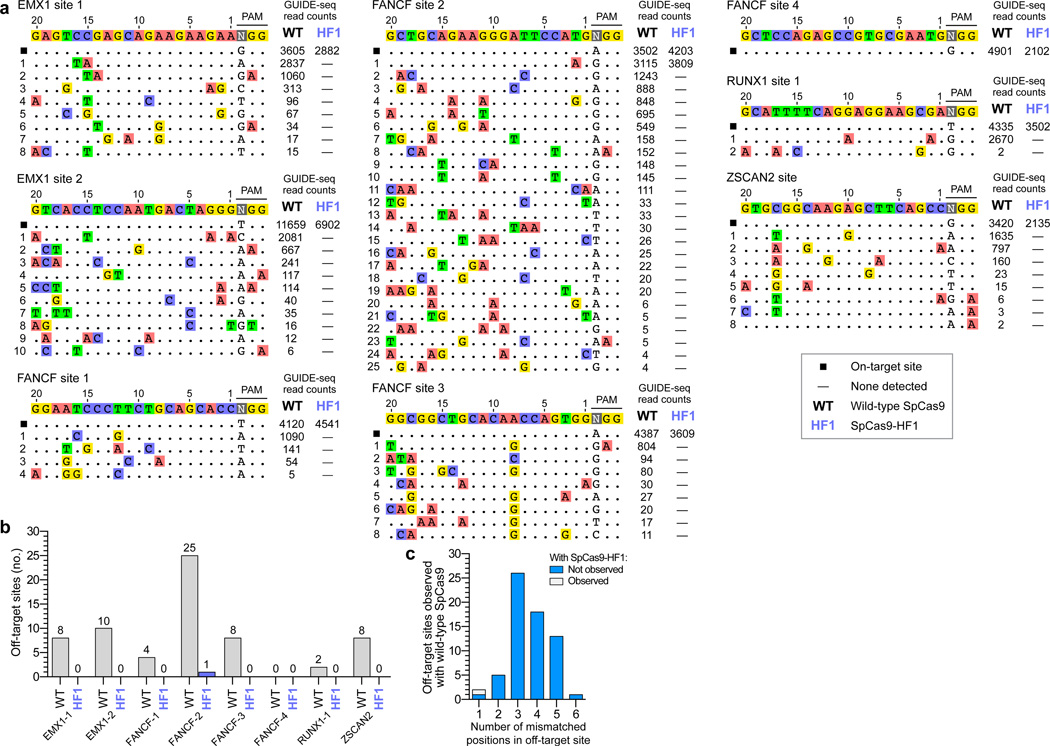
Figure 2. Genome-wide specificities of wild-type SpCas9 and SpCas9-HF1 with sgRNAs targeted to standard, non-repetitive sites.
a, Off-target cleavage sites of wild-type SpCas9 and SpCas9-HF1 with eight sgRNAs targeted to endogenous human genes, as determined by GUIDE-seq. Read counts represent a measure of cleavage frequency at a given site; mismatched positions within the spacer or PAM are highlighted in color. b, Summary of the total number of genome-wide off-target sites identified by GUIDE-seq for wild-type SpCas9 and SpCas9-HF1 with the sgRNAs used in panel a. c, Off-target sites identified for wild-type SpCas9 and SpCas9-HF1 for the eight sgRNAs, binned according to the total number of mismatches (in the protospacer and PAM) relative to the on-target site.