Fig. 4.
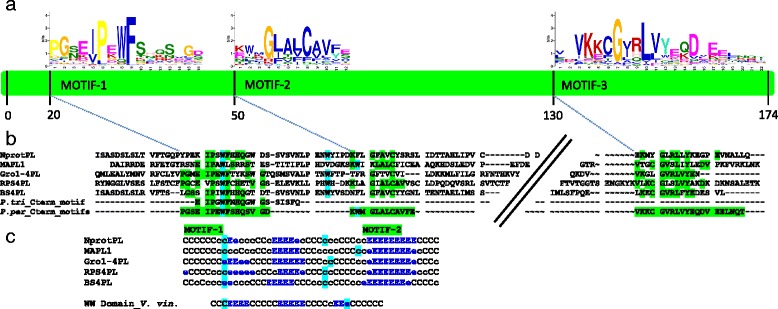
Location and structure of the three conserved motifs defined in the PL domain sequence. a The green shape outlines the position of the three motifs with their average start position. b Alignment of the five reference PL domains, the motifs described in Populus trichocarpa (P. tri_Cterm_motif) [5] and the signature found in P. persica (P. per_Cterm_motifs). Amino-acid identity is highlighted in green and essential tryptophans (W) are identified in blue. c HHpred secondary structure prediction of the 5 PL reference domains. C signifies a coil or loop residue. Predicted stands for extended beta sheet (E) are surrounded by the tryptophans (in blue). The putative WW domains (PFAM00397) display the same structural characteristics. The WW domain sequence used (ww Domain_V.vin) is extracted from the perennial species Vitis vinifera (XP_010664484.1)
