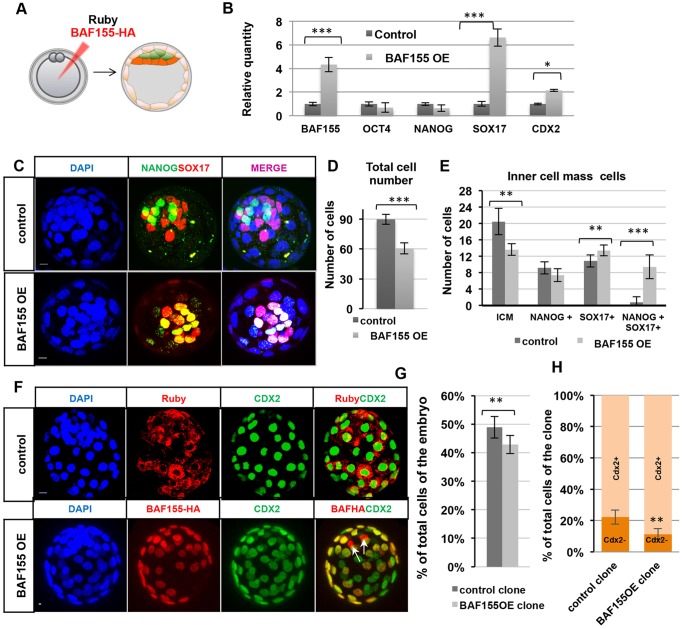
Fig. 4.
Upregulation of BAF155 shifts the developmental programme towards the extra-embryonic lineage. (A) Overexpression (OE) experiments of BAF155 using the HA-tagged BAF155 construct or of Ruby (control). (B) qRT-PCR on whole embryos, comparing lineage marker transcripts of control and BAF155 OE blastocysts. (C) Immunofluorescence images of control and BAF155 OE blastocysts at E4.5. (D) Total cell number was reduced in BAF155 OE blastocysts (61±6) compared with the control (90±5). (E) The total number of ICM cells was reduced in BAF155 OE blastocysts (14±2) compared with the control (21±3); the majority of ICM cells in BAF155 OE blastocysts co-express NANOG and SOX17 (9±3), unlike control blastocysts (1±2). (F) z-projections of control and BAF155 OE blastocysts: Ruby blastocyst contributes equally to the ICM and CDX2+ TE cell populations, whereas BAF155 OE blastocyst infrequently contributes to the CDX2− ICM cells (arrows). (G) The percentage of clones injected with BAF155 contributing to the total blastocyst was lower than that injected with Ruby. (H) Clones injected with BAF155 showed a higher contribution to the CDX2+ TE lineage compared with Ruby+ clones. Error bars represent s.d. *P<0.05, **P<0.01, ***P<0.001, Student's t-test. Scale bars: 10 µm.