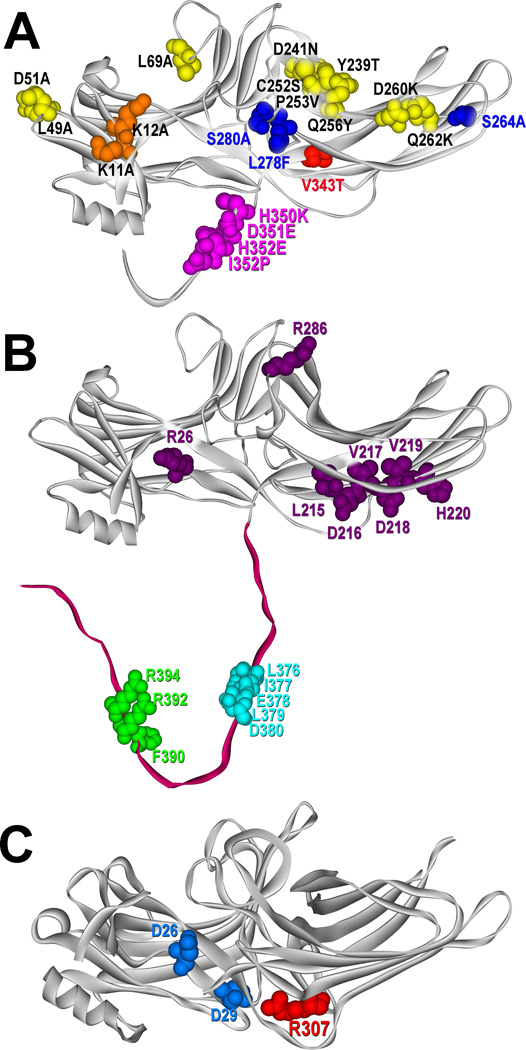
Fig. 2. Arrestin elements implicated in binding different partners.
A. The structure of arrestin-3 (PDB ID: 3P2D (Zhan et al., 2011)), showing the key lysines that bind receptor-attached phosphates (K11, K12), residues that bind other parts of receptors and determine arrestin preference for particular GPCRs (L49, D51, L69, Y239, D241, C252, P253, Q256, D260, and Q262) (Gimenez et al., 2012; Vishnivetskiy et al., 2011), and the elements responsible for arrestin-3’s ability to facilitate JNK3 activation (S264, L278, S280, V343, H350, D351, H352, and I353) (Seo et al., 2011). B. The structure of arrestin-3 (PDB ID: 3P2D (Zhan et al., 2011)), showing the key residues involved in PDE4D binding (R26, L215, D216, V217, D218, V219, H220, and R286) (Baillie et al., 2007), residues involved in clathrin binding (L376, I377, E378, L379, and D380) (Kim and Benovic, 2002; Krupnick et al., 1997), and residues engaged by the clathrin adaptor AP2 (F390, R392, and R394) (Kim and Benovic, 2002; Laporte et al., 1999). C. The structure of arrestin-2 (PDB ID: 1G4M (Han et al., 2001)), showing critical residues involved in MEK1 binding (D26 and D29) (Meng et al., 2009) and Arg-307, which is critical for cRaf1 binding (Coffa et al., 2011). All structures are rendered in ViewerPro; all highlighted residues are shown as CPK models. Parts of the C-tails not resolved in crystal structures were added manually. Color version of this figure is available online.