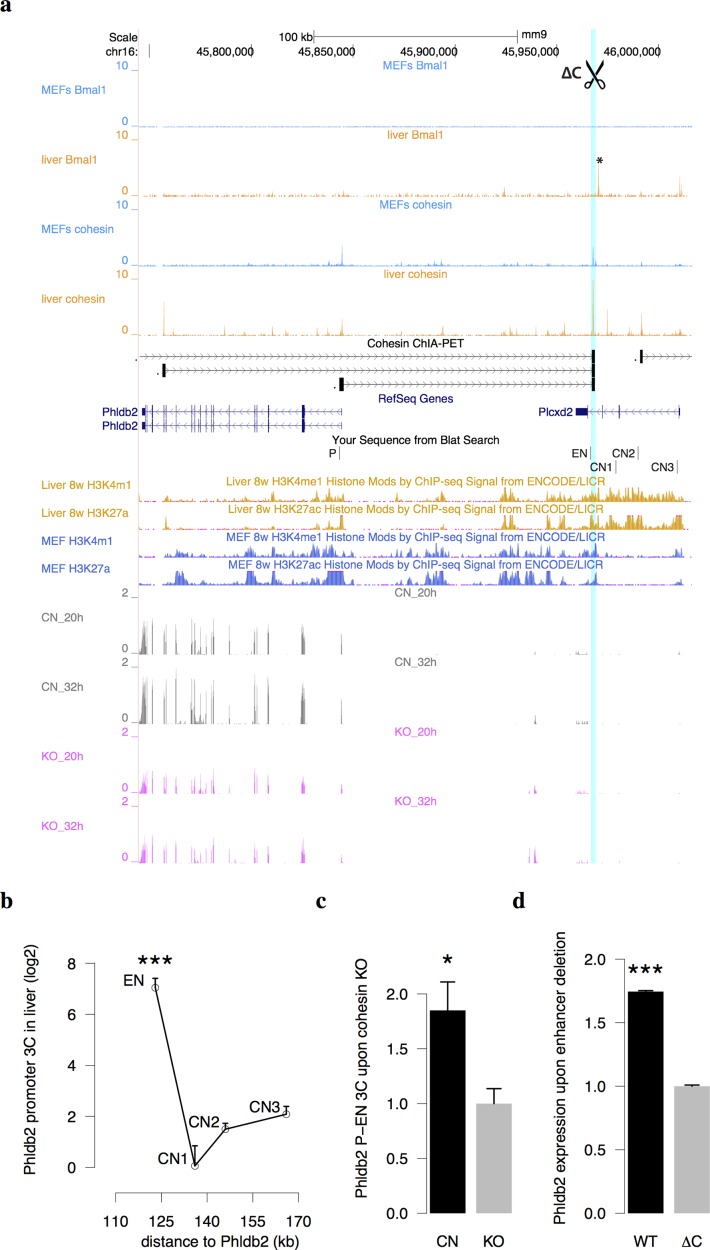
Fig 6. The expression of a COG, Phldb2, is influenced by an invariant cohesion-mediated enhancer-promoter interaction.
(a) A cohesin loop connects the BMAL1 enhancer (marked by asterisk) and the promoter of Phldb2. Conserved cohesin binding sites and active histone marks between liver and MEFs were found at the promoter of Phldb2 and its enhancer. The enhancer was bound by Bmal1 in liver but not in MEFs. The locations of primers used in 3C assay in liver and MEFs were indicated below the RefGenes. ∆C denotes CRISPR-mediated deletion region. The normalized RNA-Seq in Smc3-/- and control MEFs in this region were showed in the bottom. (b) The 3C signals of interactions anchored to Phldb2 promoter in mouse liver (ANOVA p = 10−7, mean+/-SD, 2 biological replicates, 4 technical replicates). CN, control. EN, enhancer. (c) The 3C signals of Phldb2 promoter-enhancer interaction in control and Smc3-/- MEFs (t-test p = 0.04, mean+/-SD, 2 biological replicates, 3 technical replicates). (d) The normalized expression level of Phldb2 after the deletion of cohesin binding site near the enhancer of Phldb2 in Hepa1-6 cells (t-test p = 10−11, mean+/-SD, 2 biological replicates, 3 technical replicates). ***p < 10−4, *p < 0.05.