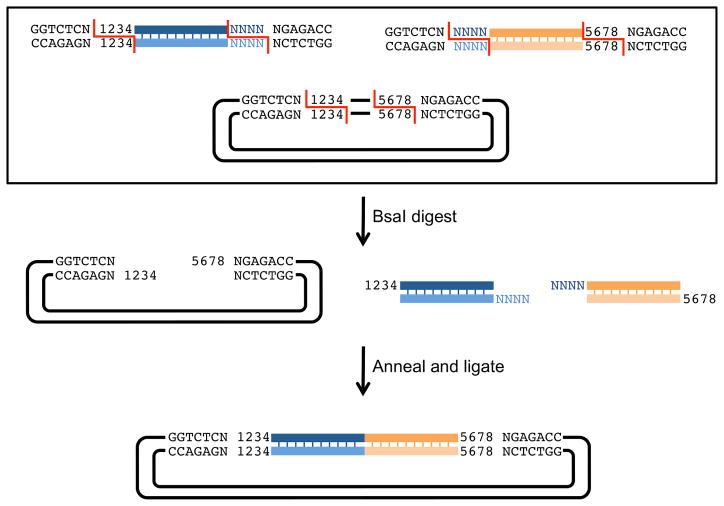
Figure 7. Multiple segment assembly using Golden Gate.
Golden Gate starting segments can be carried on separate plasmids or generated by PCR. In this example, PCR generated inserts contain flanking sequences that allow for quasi-seamless cloning of two inserts into a vector. The upstream insert has, at its 5′ end, a BsaI recognition sequence (GGTCTCN) followed by 4 nucleotides homologous to the vector (1234), and at its 3′ end, the last 4 nucleotides of the upstream insert (shown as NNNN) followed by an inverted BsaI recognition site (NGAGACC). The downstream insert has, at its 5′ end, a BsaI recognition sequence (GGTCTCN) followed by 4 nucleotides homologous to the upstream insert (NNNN), and at its 3′ end, the last 4 nucleotides of the vector (5678) followed by an inverted BsaI recognition site (NGAGACC). The vector is linearized by digestion with BsaI. After the insert segments and vector are digested, the unique 4 nucleotide overhangs anneal and are ligated by T4 ligase. The resulting plasmid has both inserts ligated without scars and flanked by BsaI recognition sites. This new assembly can be used in future constructions by the same means.