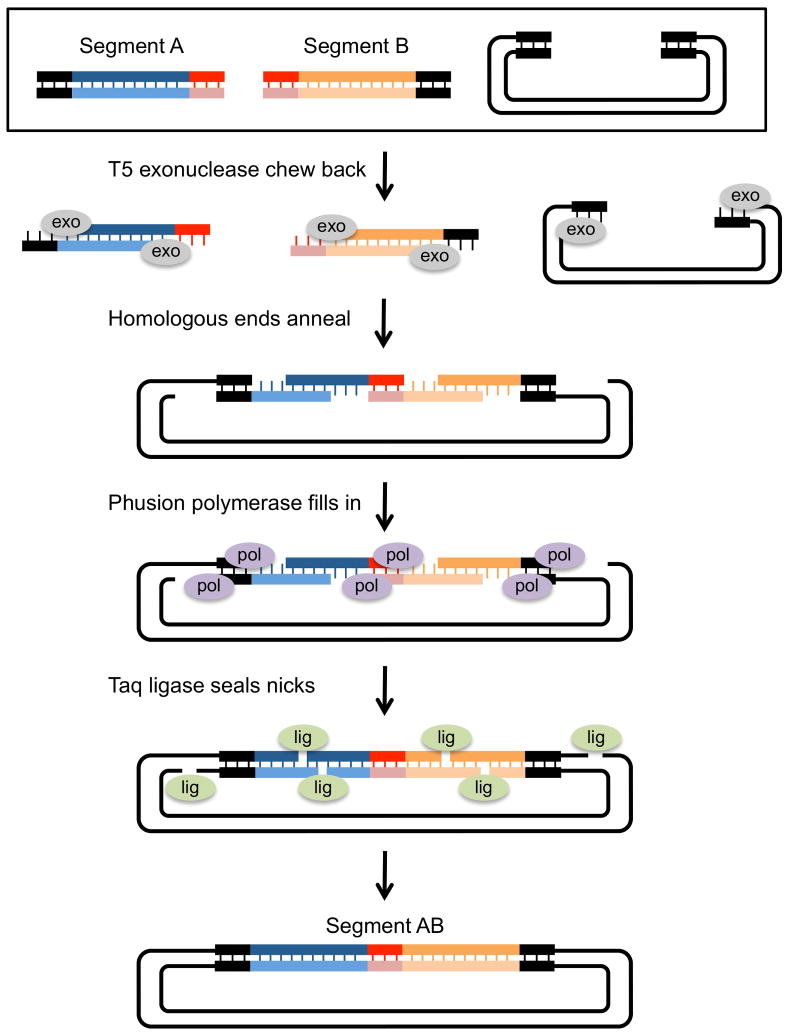
Figure 8. Multiple segment assembly using Gibson assembly.
Gibson assembly allows the seamless cloning of multiple segments into a plasmid vector. In this example, two PCR generated inserts are assembled into a linearized plasmid. Each insert segment is amplified with primers that contain sequence homology to the plasmid ends (dark ends) and to each other (light ends). The starting segments (black box) are mixed with T5 exonuclease, Phusion DNA polymerase and Taq ligase and incubated at 50 degrees for 15min to 1 hour. T5 exo chews back the 5′ ends of each segment, exposing ss 3′ homologous ends. After the 3′ ends anneal, Phusion polymerase fills in the gaps between annealed segments. In the final step, Taq ligase seals the nicks. The resulting plasmid has both inserts (Segment AB) seamlessly joined and it can be used directly as a template in PCR reactions or transformed into E coli for propagation.