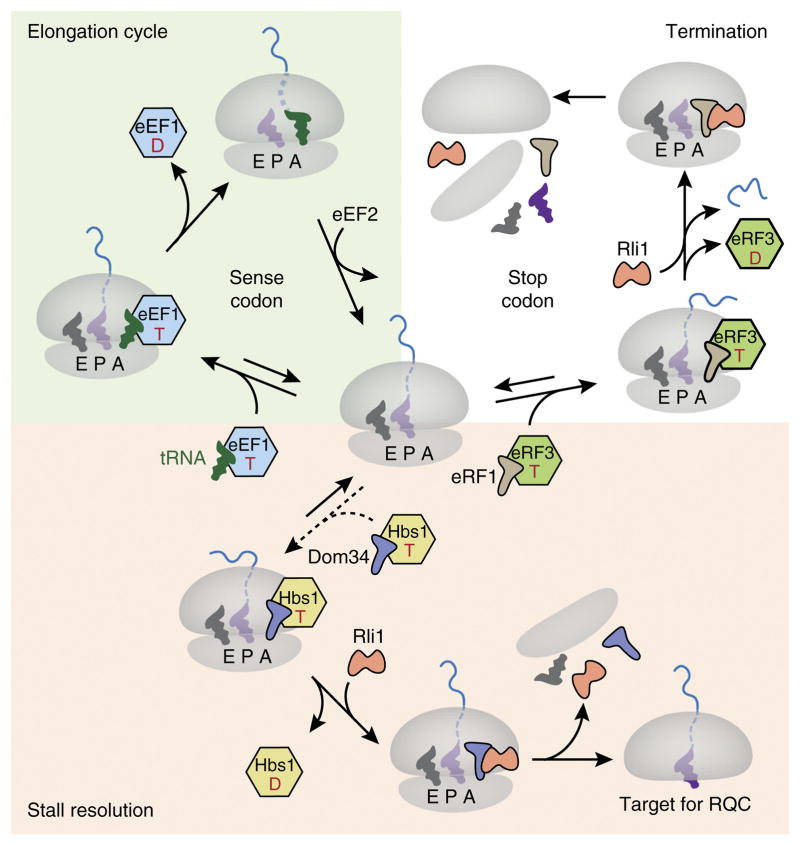
Figure 3.
Working model for recognition of a stalled ribosome by recycling factors. Top left (green background), a simplified translation elongation cycle is shown. A translating ribosome in the nonrotated state (center) engages the tRNA–eEF1A–GTP ternary complex in response to a sense codon in the A site. Codon recognition by the tRNA triggers GTP hydrolysis by eEF1A, release of the latter from the ribosome and accommodation of the tRNA to catalyze peptide-bond formation. The ribosome is then translocated by one codon via the action of eEF2 to complete the cycle. Top right (white background), when a stop codon enters the A site, it is recognized by an eRF1–eRF3–GTP complex that functions analogously to the elongation complex. Upon accommodation of eRF1, the ATPase Rli1 (ABCE1 in mammals) is recruited, and peptidyl-tRNA is hydrolyzed, thus releasing the nascent protein. The ribosomal subunits are separated by the action of the eRF1–Rli1 complex. Bottom (pink background), failure to be engaged by either the eEF1 or eRF1 complex permits ‘default’ engagement by the Dom34–Hbs1–GTP complex, which does not exhibit codon specificity. These factors act similarly to the homologous eRF1–eRF3 complex, with the exception that Dom34 (Pelota in mammals) does not catalyze peptidyl-tRNA hydrolysis. Thus, subunit separation results in a 60S–peptidyl-tRNA complex that is targeted by the RQC. T, GTP; D, GDP; E, exit tunnel.