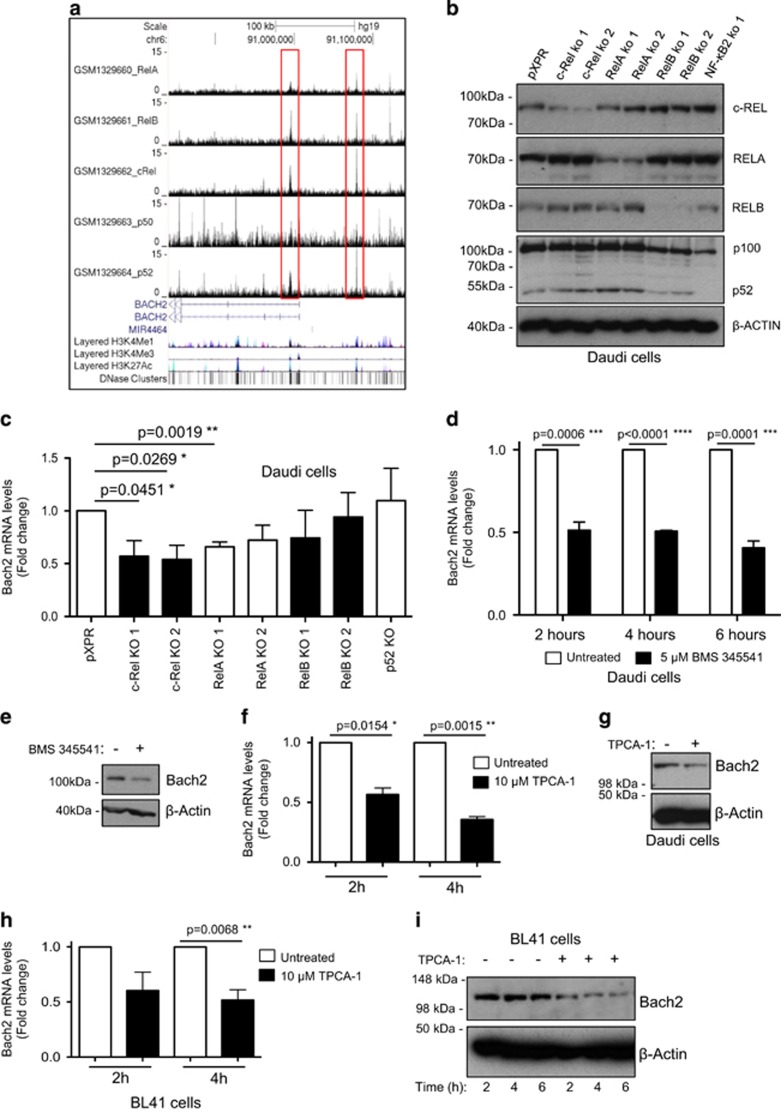
Figure 4.
Bach2 is an NF-κB target gene on human B-cell malignancies. (a) ChIP-Seq data showing NF-κB subunit binding in the region of the human BACH2 gene in the Epstein–Barr-virus-transformed lymphoblastoid B-cell line (LCL) GM12878. ChIP-Seq data were extracted from a previously published analysis of the Epstein–Barr-virus-transformed LCL GM12878 using validated anti-RelA, RelB, c-Rel, p52 and p50 antibodies.41 Reads from all ChIP-Seq experiments were mapped to the hg19/GRCh37 build of the human genome using the UCSC genome browser. (b, c) c-Rel and RelA regulate Bach2 mRNA levels in Daudi cells. In b western blot analysis shows depletion of NF-κB subunits in the Daudi Burkitt's lymphoma cell line using CRISPR/Cas9 mutagenesis. In c q-PCR shows relative Bach2 expression in the Daudi cells with mutated NF-κB subunits. Data are obtained from separately derived pools of Daudi Cas9+ cells that express either a control single-guide RNA (sgRNA) against GFP (pXPR) or an sgRNA against the indicated NF-κB subunit. RNA or protein was extracted for either q-PCR (b) or western blot (c) analysis, as indicated. Daudi Cas9/CRISPR analysis: Daudi cells with stable Cas9 expression were derived as previously described.50 Briefly, Daudi cells with stable Streptococcus pyogenes Cas9 expression were established by infection with lentiviral transduction and blasticidin selection, using pLentiCas9-Blast (Addgene plasmid #52962). Cas9 activity was validated by transduction of the Daudi Cas9+ cells with a test lentivirus, which encodes a GFP and a sgRNA that targets GFP.51 The PXPR-011 plasmid (provided by John Doench, Broad Institute, Cambridge, MA, USA) was used to construct this test virus. Cas9 activity was evident in >85% of the selected Daudi cells by flow cytometry analysis (the residual 15% of cells that continue to express GFP may be cells where the non-homologous end-joining pathway correctly repaired the Cas9-induced DNA double-strand break).51 To obtain NF-κB subunit knockdown by CRISPR/Cas9 genome editing, the following sgRNAs were designed using CRISPRdirect (http://crispr.dbcls.jp/):52 RelA 5′-AGTCCTTTCCTACAAGCTCG-3′ and 5′-AGCTGATGTGCACCGACAAG-3′ RelB 5′-GGTCTGGCGACGCGGCGACT-3′ and 5′-AGCGGCCCTCGCACTCGTAG-3′ c-Rel 5′-AAATGTGAAGGGCGATCAGC-3′ and 5′-ATTGGGTTCGAGACAACAGG-3′ p52 5′-TAGGCTGTTCCACGATCACC-3′. Oligonucleotides were synthesized by Life Technologies (Paisley, UK), were individually cloned into the lentiGuide-Puro vector (Addgene plasmid #52963), according to the protocol from the Zhang laboratory (http://genome-engineering.org/),53 and were sequence verified. VSV-G pseudotyped lentiviruses encoding a sgRNA were produced in 293 T cells and used to transduce Daudi Cas9+ cells. Cells transduced with sgRNA-encoding lentivirus were selected by puromycin. (d–g) Treatment of the Daudi Burkitt's lymphoma cell line with the IκB kinase inhibitors BMS 345541 and TPCA-1 reduces BACH2 mRNA and protein levels. Daudi cells were treated with either 5 μM BMS 345541 (Calbiochem, San Diego, CA, USA) or 10 μM TPCA-1 (Calbiochem) for the times shown. RNA or protein was extracted for either q-PCR (d, f) or western blot (e, g) analysis using the Bach2 antibody, ABN171 (Merck Millipore). (h, i) Treatment of the BL41 Burkitt's lymphoma cell lines with the IκB kinase inhibitor TPCA-1 reduces BACH2 mRNA and protein levels. BL41 cells were treated with 10μM TPCA-1 for the times shown. RNA or protein was extracted for either q-PCR (h) or western blot (i) analysis. q-PCR data represent the mean of three independent experiments±s.e.m., *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (unpaired Student's t-test). Daudi and BL41 cells were obtained from the American Type Culture Collection (Teddington, UK) and grown in RPMI-1640 medium (Lonza, Basel, Switzerland; supplemented with 10% (v/v) fetal bovine serum (Invitrogen, Paisley, UK) and 2 mM l-glutamine (Lonza)). Cell lines were sent to LGC Standards for authentication by short tandem repeat profiling.