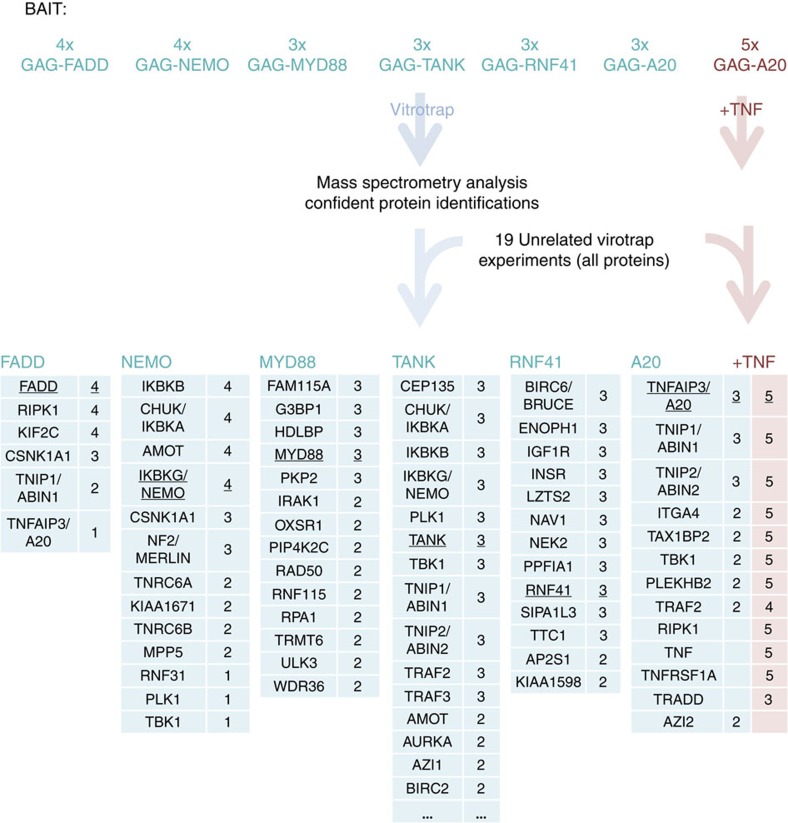
Figure 3. Use of Virotrap for unbiased interactome analysis.
A total of three (A20, TANK, MYD88 and RNF41) or four (FADD and NEMO) transfections were performed for interactome profiling. After single-step purification, specific elution, lysis and protein digestion, samples were analysed by liquid chromatography–tandem mass spectrometry. The obtained data were challenged with all the identifications obtained for 19 unrelated Virotrap experiments. The tables show the candidate interaction partners for the different baits identified with at least two peptides. The number of protein identifications in the biological repeats for the different baits is shown next to the gene name identifier. Higher recurrence is expected to increase confidence. Proteins in bold were described before (BioGRID3.2). Analysis of the A20 interactome after activation of the TNF pathway is shown as one of the conditions (in red font). Five transfections were performed for this condition.