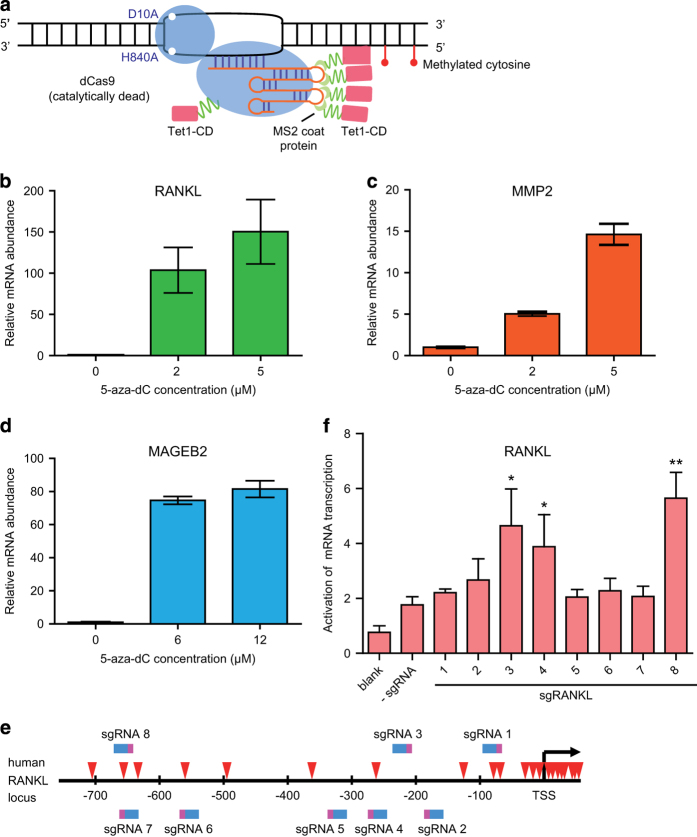
Figure 1.
CRISPR-Cas-based targeted DNA demethylation system design. (a) Schematic description of targeted demethylation via sgRNA2.0-guided recruitment of dCas9- and MS2-fused Tet1-CD. (b–d) 5-Aza-dC treatment upregulated mRNA transcription of several genes as assayed by quantitative real-time PCR (qRT-PCR) in different cells: RANKL (b) and MMP2 (c) in HEK-293FT cells, and MAGEB2 (d) in HeLa cells. The respective 5-aza-dC concentration was labeled under the columns. (e) Eight sgRNAs were selected targeting regions within −800-bp upstream to the transcription start site (TSS) of human RANKL gene. The sgRNAs recognizing their respective target sites were shown in blue-pink color (pink color represented PAM region), with the CpG sites indicated with red arrowheads. (f) RANKL mRNA expression was assayed 4 days after co-transfection of sgRANKL-(1–8) guided dCas9- and MS2-Tet1-CD in HEK-293FT using qRT-PCR assay. Results were shown after normalization to the blank control (means±s.e.m., n=3). *P<0.05, **P<0.01 compared with -sgRNA group by unpaired t-test.