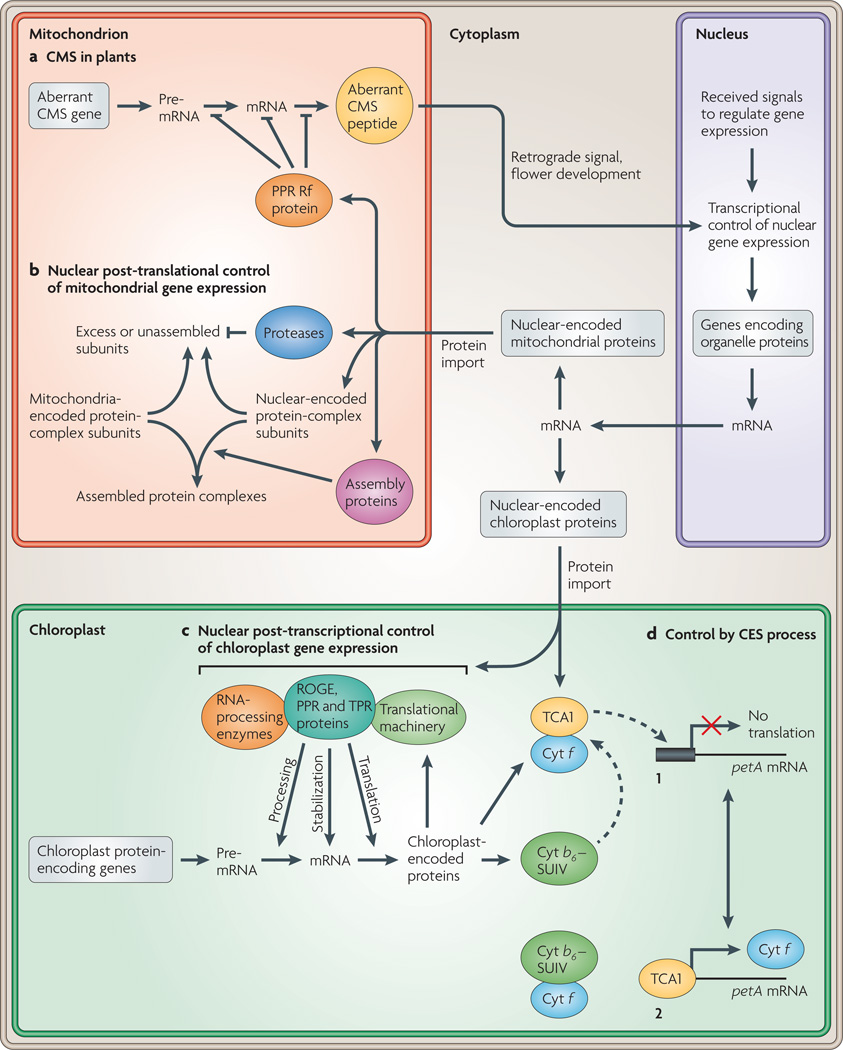
Figure 2. Nuclear anterograde control of organelle gene expression.
A generalized model of the coordination of organelle gene expression through nuclear anterograde control in eukaryotes. Several processes are highlighted. a | In flowering plants, restorers of fertility (Rf) proteins reverse the cytoplasmic male sterility (CMS) phenotype by reducing the expression of the aberrant mitochondrial gene using post-transcriptional mechanisms, b | Post-translational control of mitochondrial gene expression using nuclear-encoded proteases and assembly proteins, c | Post-transcriptional control of chloroplast gene expression using nuclear-encoded regulators of organelle gene expression (ROGE) proteins that target specific RNA transcripts, d | Control by epistasy of synthesis (CES) is an autoregulatory process in which unassembled organelle proteins repress their own translation. The autoregulation of the petA gene encoding the cytochrome f subunit (Cyt f) of the cytochrome b6f complex in Chlamydomonas reinhardtii is used as an example of the CES process. In scenario 1, unassembled Cyt f binds the ROGE protein translation factor TCA1, and translation of the petA mRNA is inhibited. In scenario 2, the presence of cognate protein subunits cytochrome b6 (Cyt b6) and cytochrome subunit IV (SUIV) assemble with Cyt f. TCA1 is now able to bind petA mRNA and activate its translation. A discussion of which proteins are organelle encoded can be found in BOX 1. A more complete model of retrograde signalling than that highlighted in panel a can be viewed in FIG. 3. Proteins and protein complexes are designated as ovals. PPR pentatricopeptide repeat; TPR, tetratricopeptide repeat.