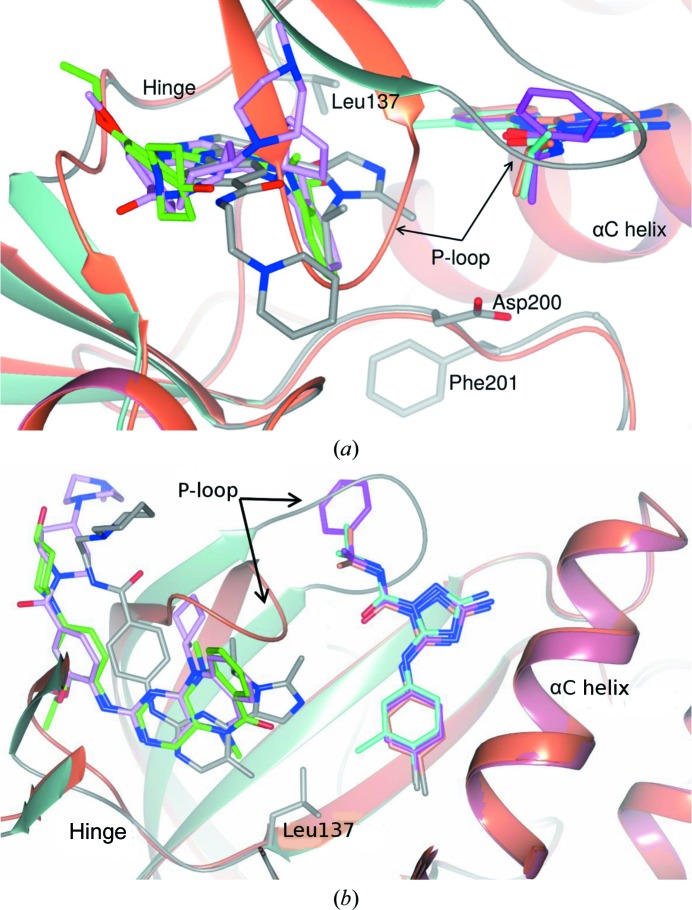
Figure 3.
Comparison of the co-crystal structures of ERK5 in complex with the conventional ATP-binding site inhibitors 1 (PDB entry 4b99), 2 and 3 with those of ERK5 in complex with the allosteric inhibitors 4, 5 and 6. Displacement of the P-loop into the ATP-binding site upon binding of 4, 5 and 6 is highlighted. The protein backbone for the complex of ERK5 with compounds 1 (coloured by secondary-structural element with β-sheets, α-helices and loops in sea green, pale crimson and grey, respectively) and 4 (coral) is shown as a worm representation. Bound ligands are rendered as sticks with C atoms coloured pink (compound 1), green (compound 2), grey (compound 3), coral (compound 4), cyan (compound 5) and magenta (compound 6). The side chains of the gatekeeper residue (Leu137) and the DFG motif (Asp200 and Phe201) from the complex of ERK5 with compound 1 are shown as sticks with C atoms in grey. In (b), the view has been rotated ∼90° with respect to (a), such that the viewer looks onto the kinase N-lobe, and the C-lobe has been omitted for clarity.