Table 1. Crystallographic statistics.
Values in parentheses are for the outer resolution shell.
| Compound | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|
| PDB code | 5byy | 5byz | 4zsg | 4zsj | 4zsl |
| Beamline | X06SA, SLS | X06SA, SLS | ID23.1, ESRF | I02, DLS | MX2, AS |
| Wavelength (Å) | 1.00003 | 1.00003 | 0.972 | 0.979 | 0.950 |
| Detector | Pilatus 6M | Pilatus 6M | ADSC Q315 CCD | ADSC Q315 CCD | ADSC Q315 CCD |
| Data-collection date | 11/06/2012 | 10/05/2012 | 22/11/2012 | 11/07/2013 | 29/08/2013 |
| Space group | P41212 | P41212 | P41212 | P41212 | P41212 |
| Unit-cell parameters | |||||
| a = b (Å) | 92.79 | 93.40 | 92.53 | 93.04 | 92.69 |
| c (Å) | 107.30 | 116.28 | 107.48 | 110.61 | 111.04 |
| Resolution range (Å) | 70.19–2.79 (3.04–2.79) | 72.82–1.65 (1.74–1.65) | 50.0–1.79 (1.85–1.79) | 71.2–2.48 (2.54–2.48) | 47.63–2.25 (2.32–2.25) |
| Completeness (%) | 99.9 (100) | 99.5 (99.8) | 99.4 (98.7) | 99.9 (99.8) | 99.8 (98.1) |
| Unique reflections | 12125 | 62054 | 44410 | 17842 | 23699 |
| Multiplicity | 5.9 (5.7) | 7.1 (7.0) | 10.6 (10.8) | 10.8 (11.3) | 7.9 (7.7) |
| Mean I/σ(I) | 21.7 (4.0) | 24.2 (3.3) | 20.9 (1.7) | 19.1 (3.5) | 17.5 (1.9) |
| R merge † | 0.069 (0.440) | 0.046 (0.690) | 0.055 (1.257) | 0.110 (0.719) | 0.089 (1.203) |
| R value‡ (%) | 22.3 | 17.4 | 20.6 | 17.0 | 17.7 |
| R free § (%) | 27.3 | 19.2 | 23.5 | 23.0 | 21.9 |
| Non-H protein atoms | 2739 | 2960 | 2794 | 2794 | 2737 |
| Non-H ligand atoms | 35 | 34 | 20 | 21 | 25 |
| Solvent molecules | 17 | 337 | 344 | 261 | 276 |
| R.m.s. deviations from ideal values | |||||
| Bond lengths (Å) | 0.008 | 0.009 | 0.010 | 0.010 | 0.010 |
| Bond angles (°) | 1.07 | 1.20 | 1.02 | 1.06 | 1.00 |
| Average B value for protein (Å2) | 23.3 | 21.7 | 55.2 | 47.9 | 46.6 |
| Average B value for ligand (Å2) | 62.0 | 23.9 | 39.2 | 68.2 | 73.7 |
| Average B value for water (Å2) | 14.5 | 30.3 | 57.6 | 56.6 | 56.4 |
| φ, ψ angle distribution for residues¶ | |||||
| In favoured regions (%) | 95 | 95.7 | 94.3 | 95.3 | 95.9 |
| In allowed regions (%) | 4.2 | 3.6 | 4.8 | 4.1 | 3.8 |
| In outlier regions (%) | 0.8 | 0.7 | 0.9 | 0.6 | 0.3 |
R
merge = 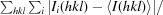
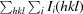 .
.
R = 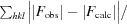
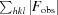 .
.
R free is the cross-validation R factor computed for a test set consisting of 5% of the unique reflections.
Ramachandran statistics as defined by Coot.
