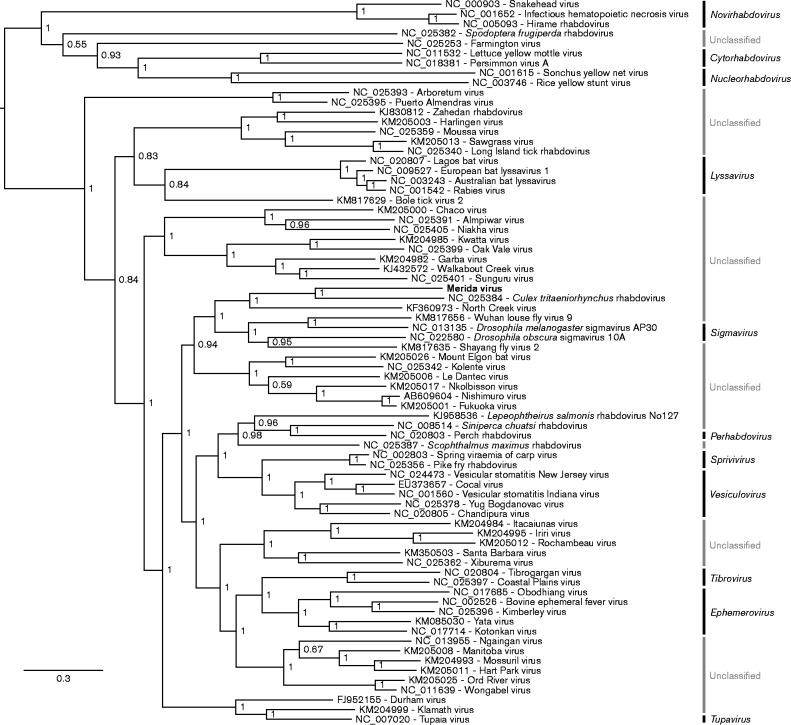
Fig. 2.
Phylogenetic tree for MERDV and selected other rhabdovirus sequences. L protein amino acid sequences were aligned using muscle (Edgar, 2004). A maximum-likelihood phylogenetic tree was estimated using the Bayesian Markov chain Monte Carlo method implemented in MrBayes version 3.2.3 (Ronquist et al., 2012) sampling across the default set of fixed amino acid rate matrices with 10 million generations, discarding the first 25 % as burn-in. The original figure was produced using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The tree is midpoint-rooted and selected nodes are labelled with posterior probability values. Rhabdovirus genera, where defined, are labelled on the far right. GenBank accession numbers are indicated next to virus names. Bar indicates amino acid substitutions per site.