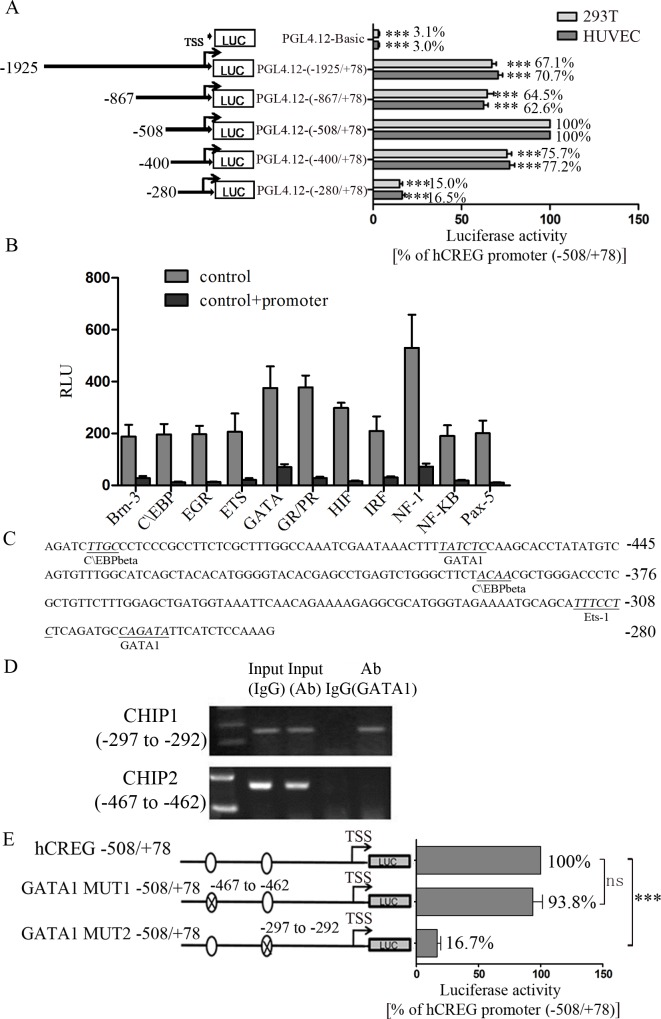
Fig 4. Identification of the core promoter of human CREG gene (hCREG) and the transcription factors that bind to the hCREG promoter.
(A) Identification of the core promoter of hCREG. A 1.9 kb genomic DNA sequence upstream of the hCREG transcription start site (TSS) linked with Xho I/Hind III restriction sites was subcloned into pGL4.12-basic luciferase reporter vectors. A series of promoter deletions were created based on the 1.9 kb hCREG-luciferase reporter. HUVECs and 293T cells were transiently transfected with reporter vectors and harvested after 48 h. For each construct, a PGL4.73 plasmid was co-transfected to correct for differences in transfection efficiency. The corrected luciferase activity was normalized to the activity of the hCREG -508/+78 plasmids (100% activity). (B) Promoter-binding transcription-factor (TF) profiling array assay of hCREG core promoter was performed. This is a competitive binding assay performed to identify promoter-bound TFs through comparisons of the results in the presence (control + promoter) or absence (control) of the hCREG core promoter. If the hCREG promoter contains a TF binding sequence, it will display a lower chemiluminescence activity. In this study, hCREG was found to potentially bind with Brn-3, C\EBP, EGR, ETS, GATA, GR/PR, HIF, IRF, NF-1, NF-κB and Pax-5. (C) Bioinformatic predictions of the consensus binding sequences of C\EBP β, GATA1, and Ets-1. (D) ChIP assay confirmed that GATA1 bound directly to hCREG at the consensus GATA binding sequence [(-297/-292 bp) upstream from TSS]. (E) Plasmids containing the hCREG promoter (-508/+78) with 2 deletion mutations at the consensus GATA1 binding element were constructed with the luciferase (Luc) gene in PGL4.12 vectors. Luciferase activity was normalized to that of the hCREG core promoter (100%). n = 9. Data are shown as the mean ± SE from 3 independent experiments performed in triplicates. *P<0.05 and ***P<0.001 compared with either the hCREG core promoter (100%) (A and E) or the control (B).